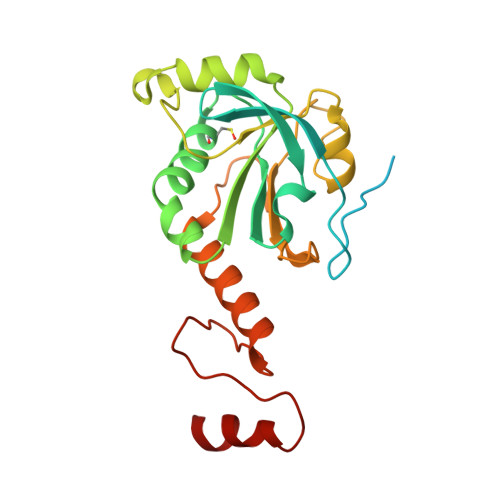
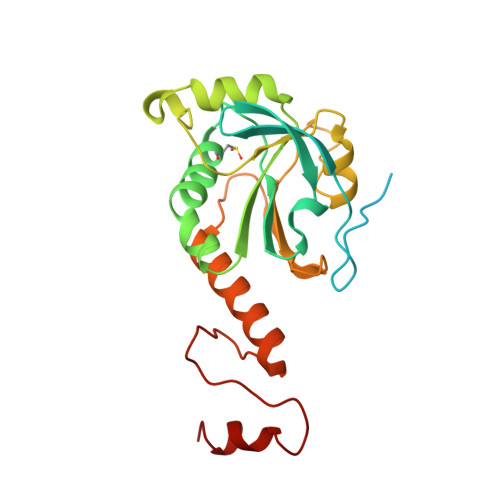
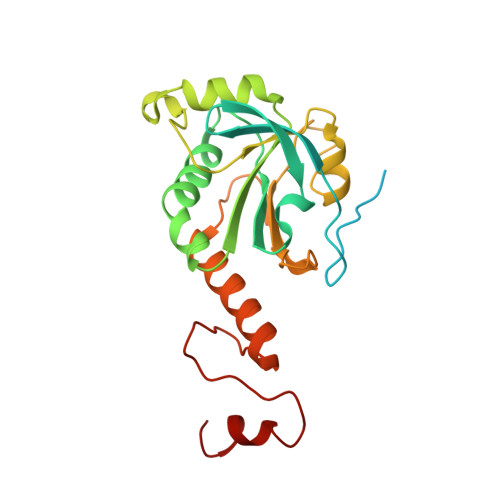
Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4
Wang, X., Wang, L., Wang, X., Sun, F., Wang, C.-C.(2011) Biochem J
- PubMed: 21916849
- DOI: https://doi.org/10.1042/BJ20110380
- Primary Citation of Related Structures:
3TKP, 3TKQ, 3TKR, 3TKS - PubMed Abstract:
Prx4 (peroxiredoxin 4) is the only peroxiredoxin located in the ER (endoplasmic reticulum) and a proposed scavenger for H2O2. In the present study, we solved crystal structures of human Prx4 in three different redox forms and characterized the reaction features of Prx4 with H2O2. Prx4 exhibits a toroid-shaped decamer constructed of five catalytic dimers. Structural analysis revealed conformational changes around helix α2 and the C-terminal reigon with a YF (Tyr-Phe) motif from the partner subunit, which are required for interchain disulfide formation between Cys87 and Cys208, a critical step of the catalysis. The structural explanation for the restricting role of the YF motif on the active site dynamics is provided in detail. Prx4 has a high reactivity with H2O2, but is susceptible to overoxidation and consequent inactivation by H2O2. Either deletion of the YF motif or dissociation into dimers decreased the susceptibility of Prx4 to overoxidation by increasing the flexibility of Cys87.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: