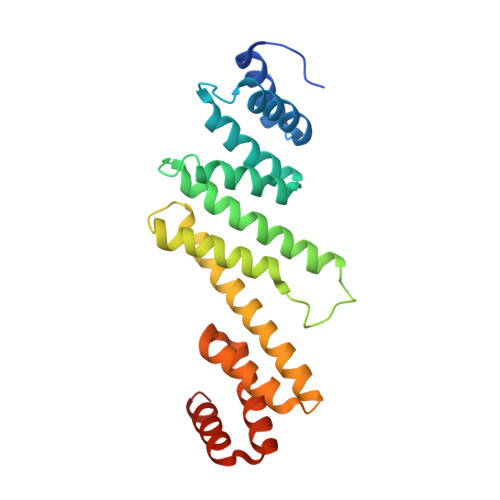
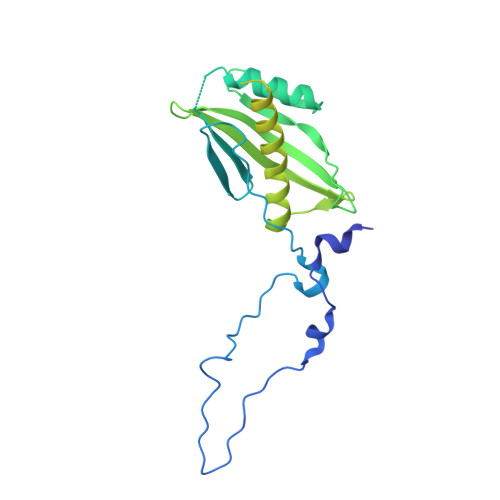
Crystal structure of the beta-barrel assembly machinery BamCD protein complex
Kim, K.H., Aulakh, S., Paetzel, M.(2011) J Biological Chem 286: 39116-39121
- PubMed: 21937441
- DOI: https://doi.org/10.1074/jbc.M111.298166
- Primary Citation of Related Structures:
3TGO - PubMed Abstract:
The β-barrel assembly machinery (BAM) complex of Escherichia coli is a multiprotein machine that catalyzes the essential process of assembling outer membrane proteins. The BAM complex consists of five proteins: one membrane protein, BamA, and four lipoproteins, BamB, BamC, BamD, and BamE. Here, we report the first crystal structure of a Bam lipoprotein complex: the essential lipoprotein BamD in complex with the N-terminal half of BamC (BamC(UN) (Asp(28)-Ala(217)), a 73-residue-long unstructured region followed by the N-terminal domain). The BamCD complex is stabilized predominantly by various hydrogen bonds and salt bridges formed between BamD and the N-terminal unstructured region of BamC. Sequence and molecular surface analyses revealed that many of the conserved residues in both proteins are found at the BamC-BamD interface. A series of truncation mutagenesis and analytical gel filtration chromatography experiments confirmed that the unstructured region of BamC is essential for stabilizing the BamCD complex structure. The unstructured N terminus of BamC interacts with the proposed substrate-binding pocket of BamD, suggesting that this region of BamC may play a regulatory role in outer membrane protein biogenesis.
- Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, British Columbia V5A 1S6, Canada.
Organizational Affiliation: