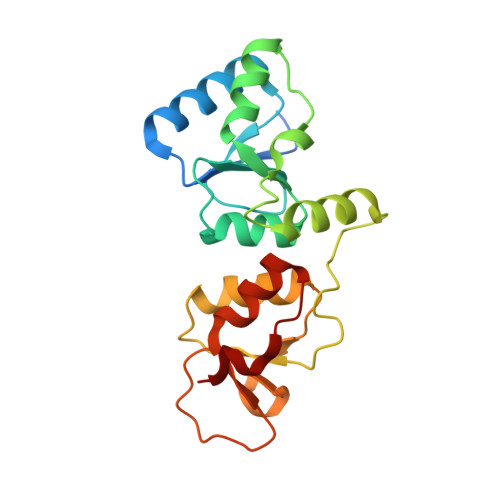
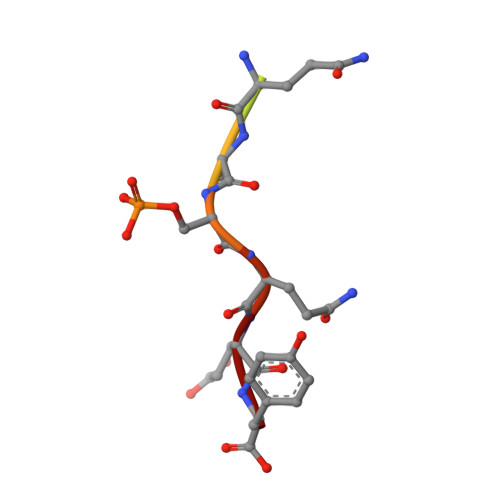
Specific recognition of phosphorylated tail of H2AX by the tandem BRCT domains of MCPH1 revealed by complex structure
Shao, Z.H., Li, F.D., Sy, S.M.-H., Yan, W., Zhang, Z., Gong, D., Wen, B., Huen, M.S.Y., Gong, Q., Wu, J., Shi, Y.(2012) J Struct Biol 177: 459-468
- PubMed: 22154951
- DOI: https://doi.org/10.1016/j.jsb.2011.11.022
- Primary Citation of Related Structures:
3SHT, 3SHV - PubMed Abstract:
MCPH1 is especially important for linking chromatin remodeling to DNA damage response. It contains three BRCT (BRCA1-carboxyl terminal) domains. The N-terminal region directly binds with chromatin remodeling complex SWI-SNF, and the C-terminal BRCT2-BRCT3 domains (tandem BRCT domains) are involved in cellular DNA damage response. The MCPH1 gene associates with evolution of brain size, and its variation can cause primary microcephaly. In this study we solve the crystal structures of MCPH1 natural variant (A761) C-terminal tandem BRCT domains alone as well as in complex with γH2AX tail. Compared with other structures of tandem BRCT domains, the most significant differences lie in phosphopeptide binding pocket. Additionally, fluorescence polarization assays demonstrate that MCPH1 tandem BRCT domains show a binding selectivity on pSer +3 and prefer to bind phosphopeptide with free COOH-terminus. Taken together, our research provides new structural insights into BRCT-phosphopeptide recognition mechanism.
- Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, PR China.
Organizational Affiliation: