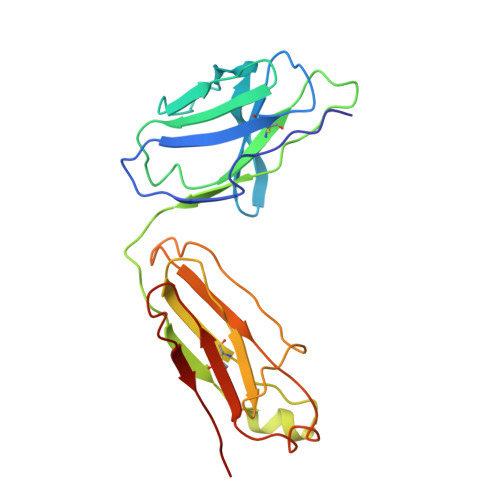
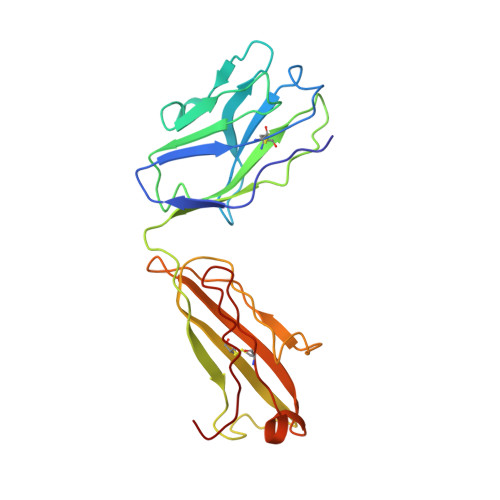
Structure of Fab fragment of malaria transmission blocking antibody 2A8 against P. vivax P25 protein
Saxena, A.K.(2012) Int J Biol Macromol 50: 153-156
- PubMed: 22037467
- DOI: https://doi.org/10.1016/j.ijbiomac.2011.10.012
- Primary Citation of Related Structures:
3S62 - PubMed Abstract:
Understanding the structural basis of recognition between antigen and antibody requires the structural comparison of free and complexed components. Previously, we have reported the crystal structure of the complex between Fab fragment of murine monoclonal antibody 2A8 (Fab2A8) and Plasmodium vivax P25 protein (Pvs25) at 3.2 Å resolution. We report here the crystallization and X-ray structure of native Fab2A8 at 4.0 Å resolution. The 2A8 antibody generated against Pvs25 prevents the formation of P. vivax oocysts in the mosquito, when assayed in membrane feeding experiment. Comparison of native Fab2A8 structure with antigen bound Fab2A8 structure indicates the significant conformational changes in CDR-H1 and CDR-H3 regions of V(H) domain and CDR-L3 region of V(L) domain of Fab2A8. Upon complex formation, the relative orientation between V(L) and V(H) domains of Fab2A8 is conserved, while significant differences are observed in elbow angles of heavy and light chains. The combing site residues of complexed Fab2A8 exhibited the reduced temperature factor compared to native Fab2A8, suggesting a loss of conformational entropy upon antigen binding.
- School of Life Sciences, Jawaharlal Nehru University, New Delhi, India. ajaysaxena@mail.jnu.ac.in
Organizational Affiliation: