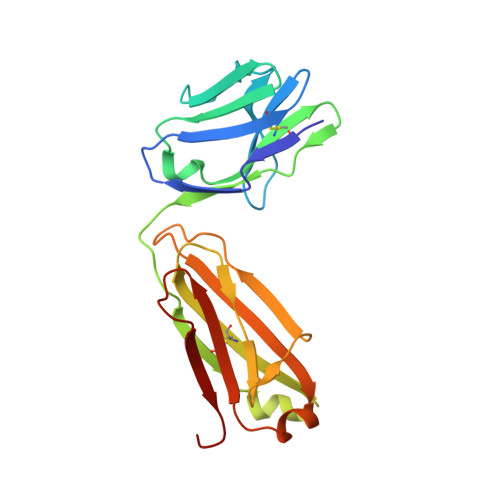
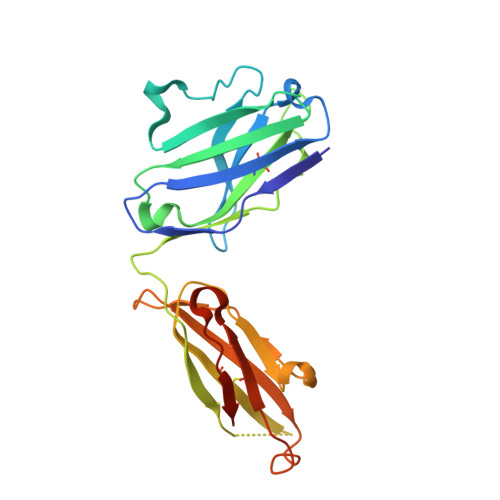
Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Scheid, J.F., Mouquet, H., Ueberheide, B., Diskin, R., Klein, F., Oliveira, T.Y., Pietzsch, J., Fenyo, D., Abadir, A., Velinzon, K., Hurley, A., Myung, S., Boulad, F., Poignard, P., Burton, D.R., Pereyra, F., Ho, D.D., Walker, B.D., Seaman, M.S., Bjorkman, P.J., Chait, B.T., Nussenzweig, M.C.(2011) Science 333: 1633-1637
- PubMed: 21764753
- DOI: https://doi.org/10.1126/science.1207227
- Primary Citation of Related Structures:
3RPI - PubMed Abstract:
Passive transfer of broadly neutralizing HIV antibodies can prevent infection, which suggests that vaccines that elicit such antibodies would be protective. Thus far, however, few broadly neutralizing HIV antibodies that occur naturally have been characterized. To determine whether these antibodies are part of a larger group of related molecules, we cloned 576 new HIV antibodies from four unrelated individuals. All four individuals produced expanded clones of potent broadly neutralizing CD4-binding-site antibodies that mimic binding to CD4. Despite extensive hypermutation, the new antibodies shared a consensus sequence of 68 immunoglobulin H (IgH) chain amino acids and arise independently from two related IgH genes. Comparison of the crystal structure of one of the antibodies to the broadly neutralizing antibody VRC01 revealed conservation of the contacts to the HIV spike.
- Laboratory of Molecular Immunology, The Rockefeller University, New York, NY 10065, USA.
Organizational Affiliation: