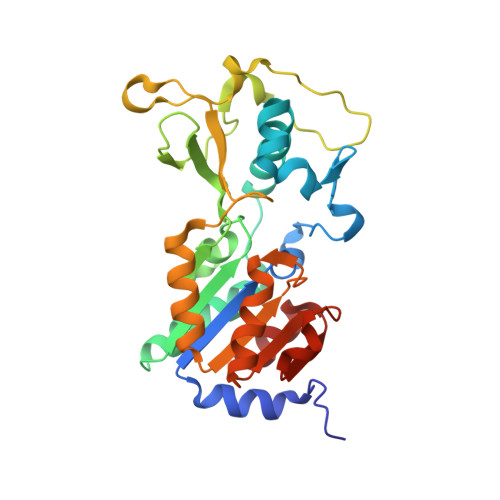
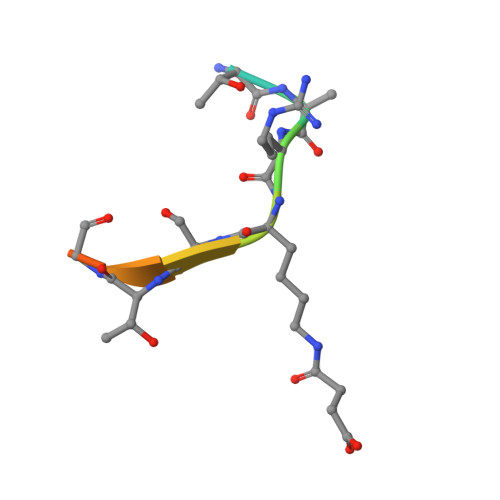
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
Du, J., Zhou, Y., Su, X., Yu, J.J., Khan, S., Jiang, H., Kim, J., Woo, J., Kim, J.H., Choi, B.H., He, B., Chen, W., Zhang, S., Cerione, R.A., Auwerx, J., Hao, Q., Lin, H.(2011) Science 334: 806-809
- PubMed: 22076378
- DOI: https://doi.org/10.1126/science.1207861
- Primary Citation of Related Structures:
3RIG, 3RIY - PubMed Abstract:
Silent information regulator 2 (Sir2) proteins (sirtuins) are nicotinamide adenine dinucleotide-dependent deacetylases that regulate important biological processes. Mammals have seven sirtuins, Sirt1 to Sirt7. Four of them (Sirt4 to Sirt7) have no detectable or very weak deacetylase activity. We found that Sirt5 is an efficient protein lysine desuccinylase and demalonylase in vitro. The preference for succinyl and malonyl groups was explained by the presence of an arginine residue (Arg(105)) and tyrosine residue (Tyr(102)) in the acyl pocket of Sirt5. Several mammalian proteins were identified with mass spectrometry to have succinyl or malonyl lysine modifications. Deletion of Sirt5 in mice appeared to increase the level of succinylation on carbamoyl phosphate synthase 1, which is a known target of Sirt5. Thus, protein lysine succinylation may represent a posttranslational modification that can be reversed by Sirt5 in vivo.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.
Organizational Affiliation: