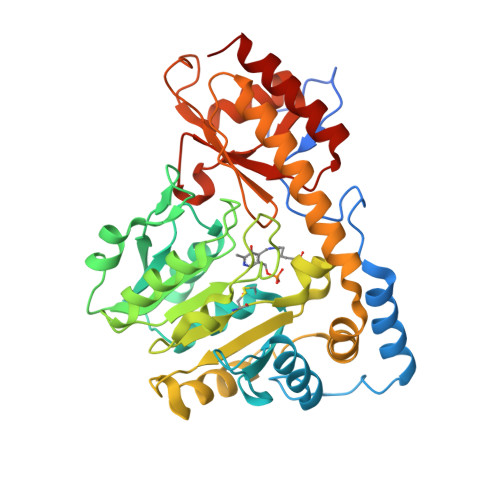
Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Mehere, P., Han, Q., Lemkul, J.A., Vavricka, C.J., Robinson, H., Bevan, D.R., Li, J.(2010) Protein Cell 1: 1023-1032
- PubMed: 21153519
- DOI: https://doi.org/10.1007/s13238-010-0128-5
- Primary Citation of Related Structures:
3PDX - PubMed Abstract:
Tyrosine aminotransferase (TAT) catalyzes the transamination of tyrosine and other aromatic amino acids. The enzyme is thought to play a role in tyrosinemia type II, hepatitis and hepatic carcinoma recovery. The objective of this study is to investigate its biochemical and structural characteristics and substrate specificity in order to provide insight regarding its involvement in these diseases. Mouse TAT (mTAT) was cloned from a mouse cDNA library, and its recombinant protein was produced using Escherichia coli cells and purified using various chromatographic techniques. The recombinant mTAT is able to catalyze the transamination of tyrosine using α-ketoglutaric acid as an amino group acceptor at neutral pH. The enzyme also can use glutamate and phenylalanine as amino group donors and p-hydroxy-phenylpyruvate, phenylpyruvate and alpha-ketocaproic acid as amino group acceptors. Through macromolecular crystallography we have determined the mTAT crystal structure at 2.9 Å resolution. The crystal structure revealed the interaction between the pyridoxal-5'-phosphate cofactor and the enzyme, as well as the formation of a disulphide bond. The detection of disulphide bond provides some rational explanation regarding previously observed TAT inactivation under oxidative conditions and reactivation of the inactive TAT in the presence of a reducing agent. Molecular dynamics simulations using the crystal structures of Trypanosoma cruzi TAT and human TAT provided further insight regarding the substrate-enzyme interactions and substrate specificity. The biochemical and structural properties of TAT and the binding of its cofactor and the substrate may help in elucidation of the mechanism of TAT inhibition and activation.
- Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Organizational Affiliation: