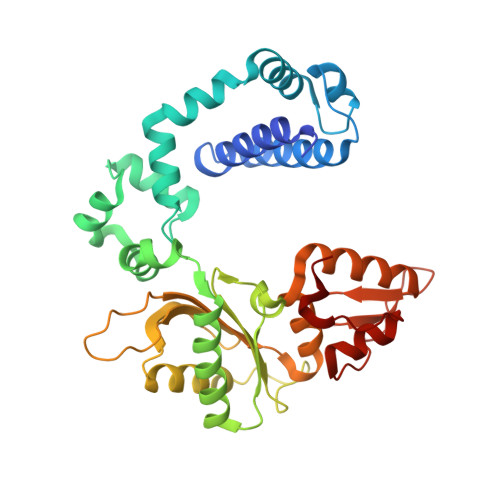
Human DNA polymerase beta mutations allowing efficient abasic site bypass.
Gieseking, S., Bergen, K., Di Pasquale, F., Diederichs, K., Welte, W., Marx, A.(2011) J Biological Chem 286: 4011-4020
- PubMed: 21107011
- DOI: https://doi.org/10.1074/jbc.M110.176826
- Primary Citation of Related Structures:
3OGU - PubMed Abstract:
The DNA of every cell in the human body gets damaged more than 50,000 times a day. The most frequent damages are abasic sites. This kind of damage blocks proceeding DNA synthesis by several DNA polymerases that are involved in DNA replication and repair. The mechanistic basis for the incapability of these DNA polymerases to bypass abasic sites is not clarified. To gain insights into the mechanistic basis, we intended to identify amino acid residues that govern for the pausing of DNA polymerase β when incorporating a nucleotide opposite to abasic sites. Human DNA polymerase β was chosen because it is a well characterized DNA polymerase and serves as model enzyme for studies of DNA polymerase mechanisms. Moreover, it acts as the main gap-filling enzyme in base excision repair, and human tumor studies suggest a link between DNA polymerase β and cancer. In this study we employed high throughput screening of a library of more than 11,000 human DNA polymerase β variants. We identified two mutants that have increased ability to incorporate a nucleotide opposite to an abasic site. We found that the substitutions E232K and T233I promote incorporation opposite the lesion. In addition to this feature, the variants have an increased activity and a lower fidelity when processing nondamaged DNA. The mutations described in this work are located in well characterized regions but have not been reported before. A crystallographic structure of one of the mutants was obtained, providing structural insights.
- Department of Chemistry, Konstanz Research School Chemical Biology, University of Konstanz, 78464 Konstanz, Germany.
Organizational Affiliation: