Structure of four mutant forms of yeast F1 ATPase: alpha-F405S
Arsenieva, D., Symersky, J., Wang, Y., Pagadala, V., Mueller, D.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit alpha | 510 | Saccharomyces cerevisiae | Mutation(s): 1 Gene Names: ATP1, YBL099W, YBL0827 EC: 3.6.3.14 | 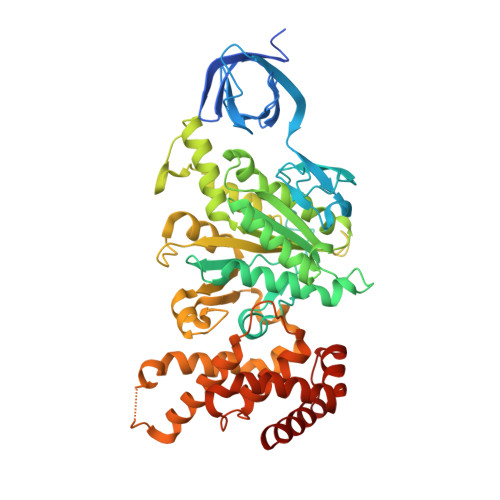 | |
UniProt | |||||
Find proteins for P07251 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P07251 Go to UniProtKB: P07251 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07251 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit beta | 484 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: ATP2, YJR121W, J2041 EC: 3.6.3.14 (PDB Primary Data), 7.1.2.2 (UniProt) | 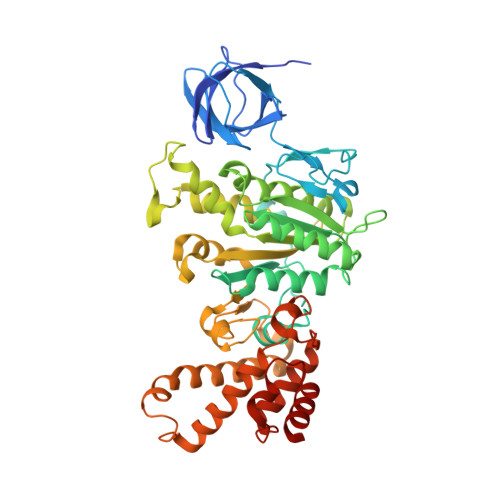 | |
UniProt | |||||
Find proteins for P00830 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P00830 Go to UniProtKB: P00830 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00830 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit gamma | 278 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: ATP3, ATP3a, ATP3b, YBR039W, YBR0408 EC: 3.6.3.14 | 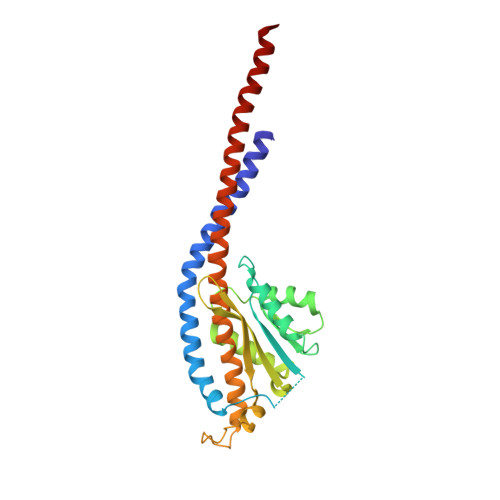 | |
UniProt | |||||
Find proteins for P38077 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P38077 Go to UniProtKB: P38077 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38077 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit delta | 138 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: ATP16, YDL004W, YD8119.03, D2935 EC: 3.6.3.14 | 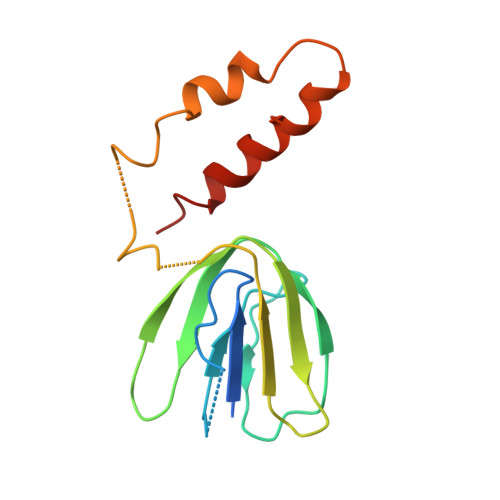 | |
UniProt | |||||
Find proteins for Q12165 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q12165 Go to UniProtKB: Q12165 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12165 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit epsilon | AA [auth 1], I, R | 61 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: ATP15, YPL271W, P0345 EC: 3.6.3.14 |  |
UniProt | |||||
Find proteins for P21306 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P21306 Go to UniProtKB: P21306 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P21306 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP Query on ANP | BA [auth A] BB [auth U] DA [auth B] DB [auth V] FA [auth C] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| PO4 Query on PO4 | JA [auth E], UA [auth N] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| MG Query on MG | AB [auth T] CA [auth A] CB [auth U] EA [auth B] EB [auth V] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 110.43 | α = 90 |
| b = 288.58 | β = 101.37 |
| c = 187.17 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |