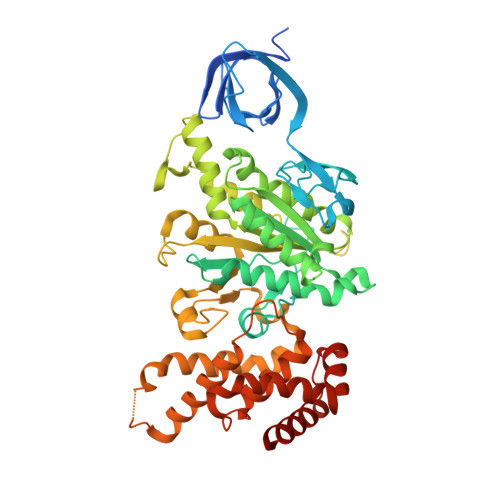
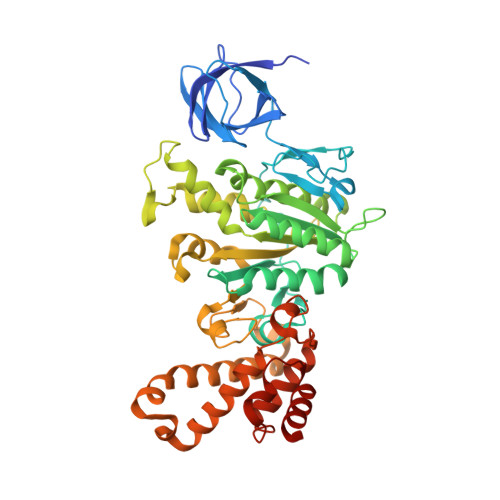
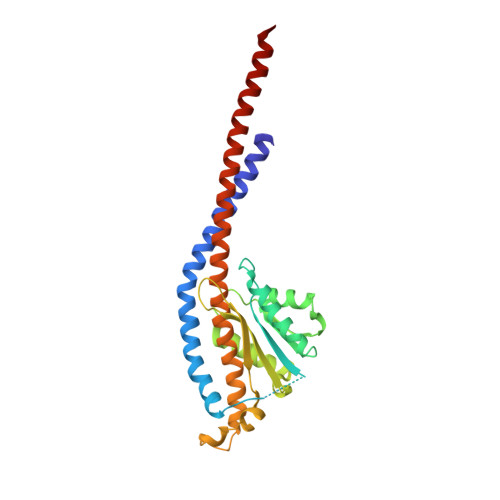
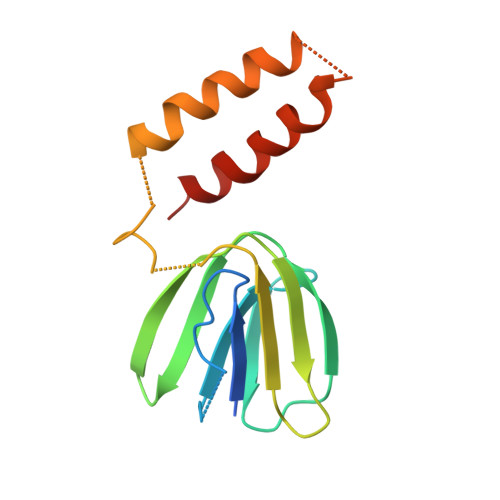
Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
Arsenieva, D., Symersky, J., Wang, Y., Pagadala, V., Mueller, D.M.(2010) J Biological Chem 285: 36561-36569
- PubMed: 20843806
- DOI: https://doi.org/10.1074/jbc.M110.174383
- Primary Citation of Related Structures:
3OE7, 3OEH, 3OFN - PubMed Abstract:
The mitochondrial ATP synthase couples the flow of protons with the phosphorylation of ADP. A class of mutations, the mitochondrial genome integrity (mgi) mutations, has been shown to uncouple this process in the yeast mitochondrial ATP synthase. Four mutant forms of the yeast F(1) ATPase with mgi mutations were crystallized; the structures were solved and analyzed. The analysis identifies two mechanisms of structural uncoupling: one in which the empty catalytic site is altered and in doing so, apparently disrupts substrate (phosphate) binding, and a second where the steric hindrance predicted between γLeu83 and β(DP) residues, Leu-391 and Glu-395, located in Catch 2 region, is reduced allowing rotation of the γ-subunit with less impedance. Overall, the structures provide key insights into the critical interactions in the yeast ATP synthase involved in the coupling process.
- Department of Biochemistry and Molecular Biology, Rosalind Franklin University of Medicine and Science, The Chicago Medical School, North Chicago, Illinois 60064, USA.
Organizational Affiliation: