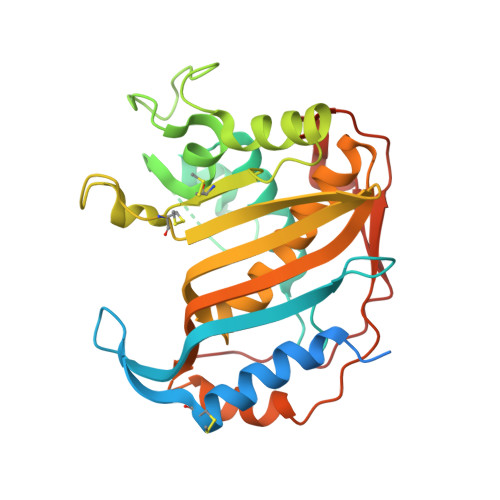
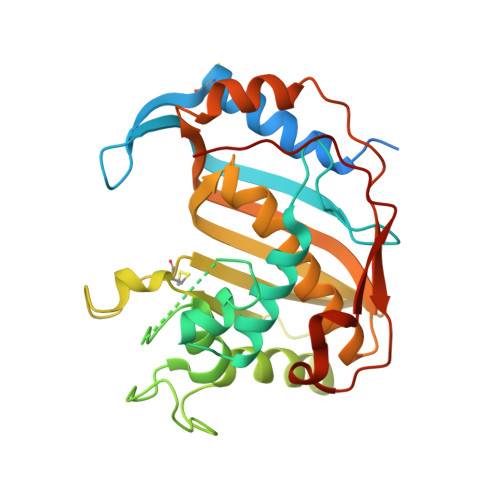
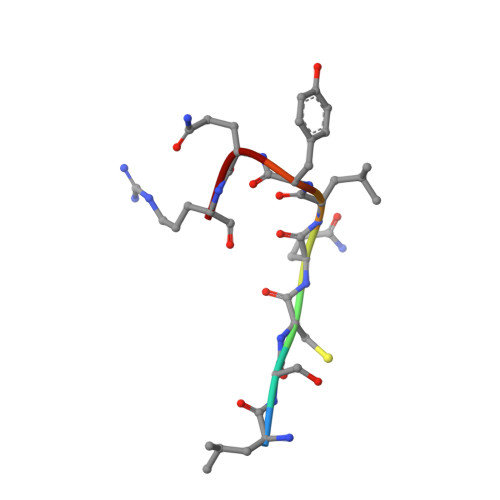
Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Cardinale, D., Guaitoli, G., Tondi, D., Luciani, R., Henrich, S., Salo-Ahen, O.M., Ferrari, S., Marverti, G., Guerrieri, D., Ligabue, A., Frassineti, C., Pozzi, C., Mangani, S., Fessas, D., Guerrini, R., Ponterini, G., Wade, R.C., Costi, M.P.(2011) Proc Natl Acad Sci U S A 108: E542-E549
- PubMed: 21795601
- DOI: https://doi.org/10.1073/pnas.1104829108
- Primary Citation of Related Structures:
3N5E, 3N5G - PubMed Abstract:
Human thymidylate synthase is a homodimeric enzyme that plays a key role in DNA synthesis and is a target for several clinically important anticancer drugs that bind to its active site. We have designed peptides to specifically target its dimer interface. Here we show through X-ray diffraction, spectroscopic, kinetic, and calorimetric evidence that the peptides do indeed bind at the interface of the dimeric protein and stabilize its di-inactive form. The "LR" peptide binds at a previously unknown binding site and shows a previously undescribed mechanism for the allosteric inhibition of a homodimeric enzyme. It inhibits the intracellular enzyme in ovarian cancer cells and reduces cellular growth at low micromolar concentrations in both cisplatin-sensitive and -resistant cells without causing protein overexpression. This peptide demonstrates the potential of allosteric inhibition of hTS for overcoming platinum drug resistance in ovarian cancer.
- Department of Pharmaceutical Sciences, via Campi 183, University of Modena and Reggio Emilia, 41126 Modena, Italy.
Organizational Affiliation: