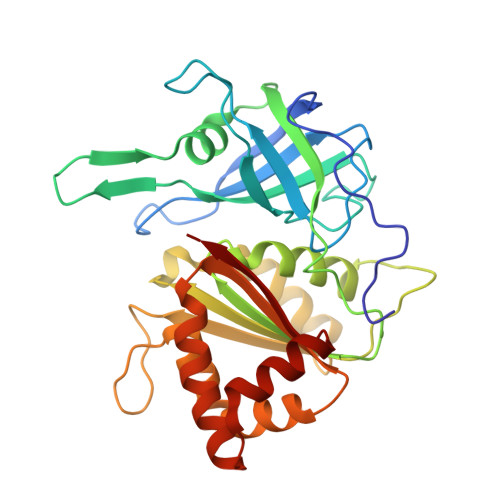
Ferredoxin:NADPH oxidoreductase is recruited to thylakoids by binding to a polyproline type II helix in a pH-dependent manner.
Alte, F., Stengel, A., Benz, J.P., Petersen, E., Soll, J., Groll, M., Bolter, B.(2010) Proc Natl Acad Sci U S A 107: 19260-19265
- PubMed: 20974920
- DOI: https://doi.org/10.1073/pnas.1009124107
- Primary Citation of Related Structures:
3MHP - PubMed Abstract:
Ferredoxin:NADPH oxidoreductase (FNR) is a key enzyme of photosynthetic electron transport required for generation of reduction equivalents. Recently, two proteins were found to be involved in membrane-anchoring of FNR by specific interaction via a conserved Ser/Pro-rich motif: Tic62 and Trol. Our crystallographic study reveals that the FNR-binding motif, which forms a polyproline type II helix, induces self-assembly of two FNR monomers into a back-to-back dimer. Because binding occurs opposite to the FNR active sites, its activity is not affected by the interaction. Surface plasmon resonance analyses disclose a high affinity of FNR to the binding motif, which is strongly increased under acidic conditions. The pH of the chloroplast stroma changes dependent on the light conditions from neutral to slightly acidic in complete darkness or to alkaline at saturating light conditions. Recruiting of FNR to the thylakoids could therefore represent a regulatory mechanism to adapt FNR availability/activity to photosynthetic electron flow.
- Munich Center for Integrated Protein Science, Ludwig-Maximilians-Universität, 81377 Munich, Germany.
Organizational Affiliation: