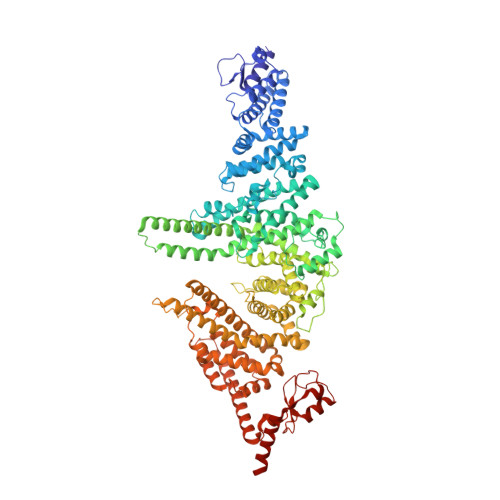
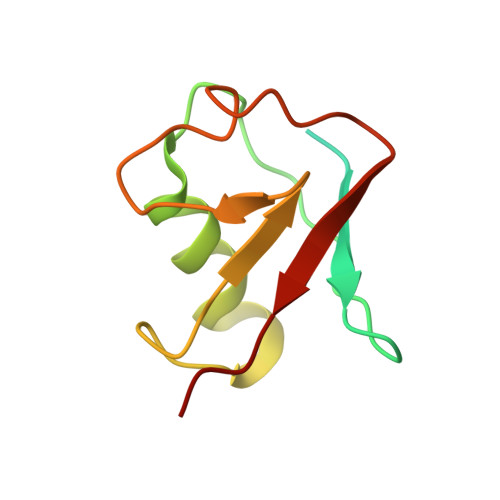
The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain.
Hanzelmann, P., Stingele, J., Hofmann, K., Schindelin, H., Raasi, S.(2010) J Biological Chem 285: 20390-20398
- PubMed: 20427284
- DOI: https://doi.org/10.1074/jbc.M110.112532
- Primary Citation of Related Structures:
3M62, 3M63 - PubMed Abstract:
Proteins containing ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains interact with various binding partners and function as hubs during ubiquitin-mediated protein degradation. A common interaction of the budding yeast UBL-UBA proteins Rad23 and Dsk2 with the E4 ubiquitin ligase Ufd2 has been described in endoplasmic reticulum-associated degradation among other pathways. The UBL domains of Rad23 and Dsk2 play a prominent role in this process by interacting with Ufd2 and different subunits of the 26 S proteasome. Here, we report crystal structures of Ufd2 in complex with the UBL domains of Rad23 and Dsk2. The N-terminal UBL-interacting region of Ufd2 exhibits a unique sequence pattern, which is distinct from any known ubiquitin- or UBL-binding domain identified so far. Residue-specific differences exist in the interactions of these UBL domains with Ufd2, which are coupled to subtle differences in their binding affinities. The molecular details of their differential interactions point to a role for adaptive evolution in shaping these interfaces.
- Rudolf Virchow Center for Experimental Biomedicine, University of Würzburg, Josef-Schneider-Strasse 2, 97080 Würzburg, Germany.
Organizational Affiliation: