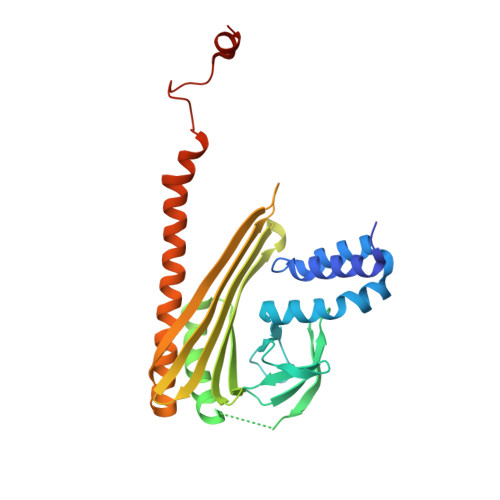
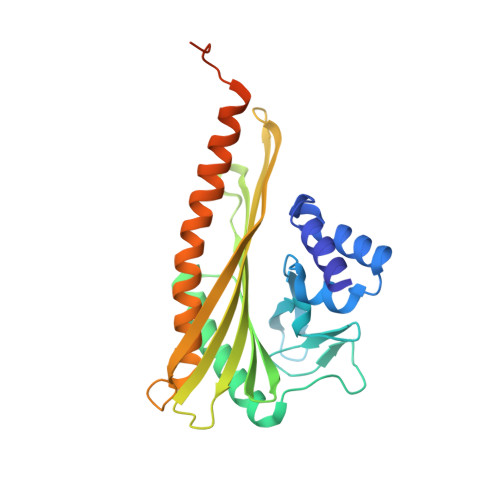
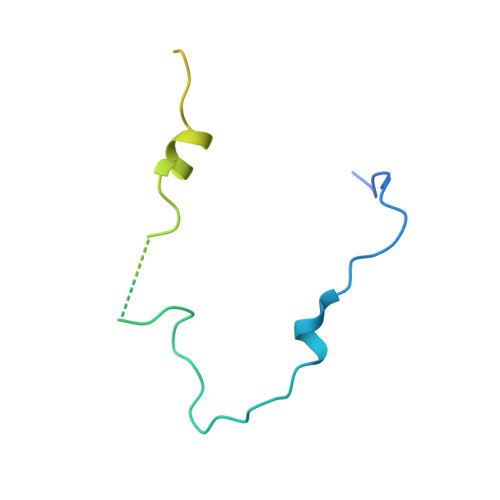
Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Hernandez-Valladares, M., Kim, T., Kannan, B., Tung, A., Aguda, A.H., Larsson, M., Cooper, J.A., Robinson, R.C.(2010) Nat Struct Mol Biol 17: 497-503
- PubMed: 20357771
- DOI: https://doi.org/10.1038/nsmb.1792
- Primary Citation of Related Structures:
3LK2, 3LK3, 3LK4 - PubMed Abstract:
Capping protein (CP) regulates actin dynamics by binding the barbed ends of actin filaments. Removal of CP may be one means to harness actin polymerization for processes such as cell movement and endocytosis. Here we structurally and biochemically investigated a CP interaction (CPI) motif present in the otherwise unrelated proteins CARMIL and CD2AP. The CPI motif wraps around the stalk of the mushroom-shaped CP at a site distant from the actin-binding interface, which lies on the top of the mushroom cap. We propose that the CPI motif may act as an allosteric modulator, restricting CP to a low-affinity, filament-binding conformation. Structure-based sequence alignments extend the CPI motif-containing family to include CIN85, CKIP-1, CapZIP and a relatively uncharacterized protein, WASHCAP (FAM21). Peptides comprising these CPI motifs are able to inhibit CP and to uncap CP-bound actin filaments.
- Institute of Molecular and Cell Biology, A*STAR, Proteos, Singapore.
Organizational Affiliation: