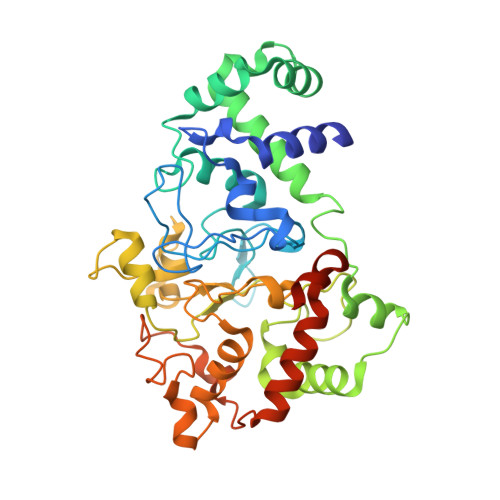
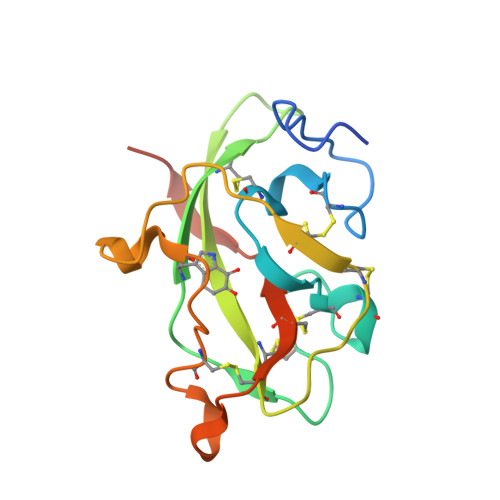
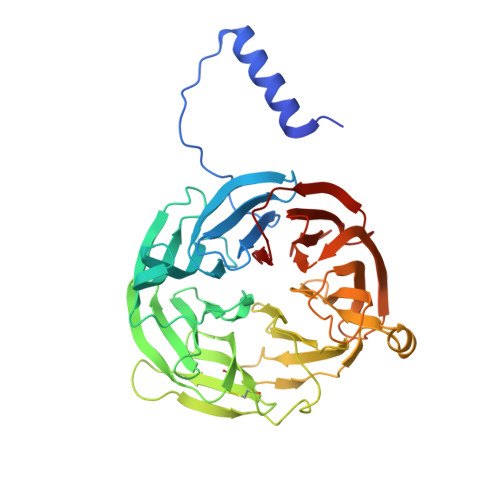
In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex.
Jensen, L.M., Sanishvili, R., Davidson, V.L., Wilmot, C.M.(2010) Science 327: 1392-1394
- PubMed: 20223990
- DOI: https://doi.org/10.1126/science.1182492
- Primary Citation of Related Structures:
3L4M, 3L4O - PubMed Abstract:
MauG is a diheme enzyme responsible for the posttranslational modification of two tryptophan residues to form the tryptophan tryptophylquinone (TTQ) cofactor of methylamine dehydrogenase (MADH). MauG converts preMADH, containing monohydroxylated betaTrp57, to fully functional MADH by catalyzing the insertion of a second oxygen atom into the indole ring and covalently linking betaTrp57 to betaTrp108. We have solved the x-ray crystal structure of MauG complexed with preMADH to 2.1 angstroms. The c-type heme irons and the nascent TTQ site are separated by long distances over which electron transfer must occur to achieve catalysis. In addition, one of the hemes has an atypical His-Tyr axial ligation. The crystalline protein complex is catalytically competent; upon addition of hydrogen peroxide, MauG-dependent TTQ synthesis occurs.
- Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, MN 55455, USA.
Organizational Affiliation: