Crystal structure of an arginine 3rd transport system periplasmic binding protein from Legionella pneumophila
Palani, K., Burley, S.K., Swaminathan, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Arginine 3rd transport system periplasmic binding protein | 237 | Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | Mutation(s): 0 Gene Names: artJ, lpg1555 | 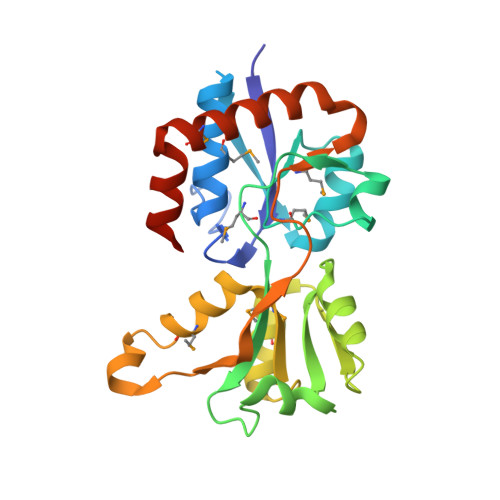 | |
UniProt | |||||
Find proteins for Q5ZV85 (Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)) Explore Q5ZV85 Go to UniProtKB: Q5ZV85 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5ZV85 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C, D | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 41.965 | α = 90 |
| b = 148.254 | β = 90 |
| c = 149.262 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| CBASS | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| SHELXD | phasing |
| SHARP | phasing |
| ARP/wARP | model building |