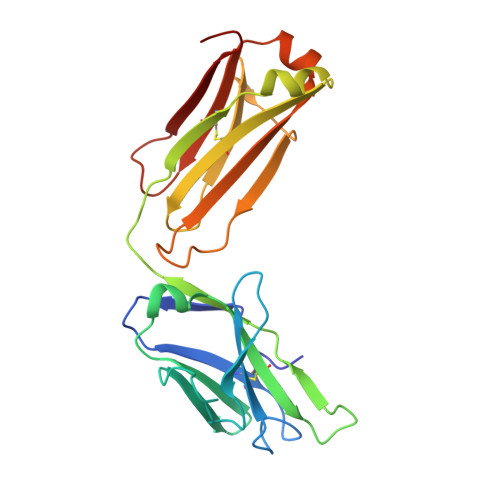
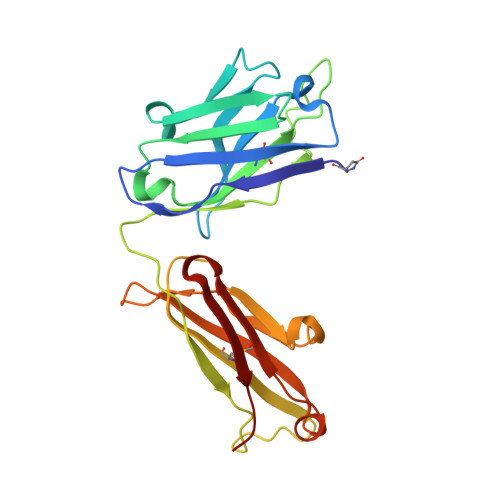
The testosterone binding mechanism of an antibody derived from a naive human scFv library
Niemi, M.H., Takkinen, K., Amundsen, L.K., Soderlund, H., Rouvinen, J., Hoyhtya, M.(2010) J Mol Recognit 24: 209-219
- PubMed: 21360611
- DOI: https://doi.org/10.1002/jmr.1039
- Primary Citation of Related Structures:
3KDM - PubMed Abstract:
A testosterone binding scFv antibody was isolated from a naïve human library with a modest size of 10(8) clones. The crystal structure of the Fab fragment form of the 5F2 antibody clone complexed with testosterone determined at 1.5 Å resolution shows that the hapten is bound deeply in the antibody binding pocket. In addition to the interactions with framework residues only CDR-L3 and CDR-H3 loops interact with testosterone and the heavy chain forms the majority of the contacts with the hapten. The testosterone binding site of the 5F2 antibody with a high abundance of aromatic amino acid residues shows similarity with an in vitro affinity matured antibody having around 300 times higher affinity. The moderate affinity of the 5F2 antibody originates from the different orientation of the hapten and few light chain contacts. This is the first three-dimensional structure of a human steroid hormone binding antibody that has been isolated from a naïve human repertoire.
- Department of Chemistry, University of Eastern Finland, P.O. Box 111, FIN-80101 Joensuu, Finland.
Organizational Affiliation: