Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
Kang, G.B., Ko, S., Song, S.M., Lee, W., Eom, S.M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquitin-conjugating enzyme E2 K | 201 | Homo sapiens | Mutation(s): 0 Gene Names: UBE2K, HIP2, LIG EC: 6.3.2.19 (PDB Primary Data), 2.3.2.23 (UniProt) | 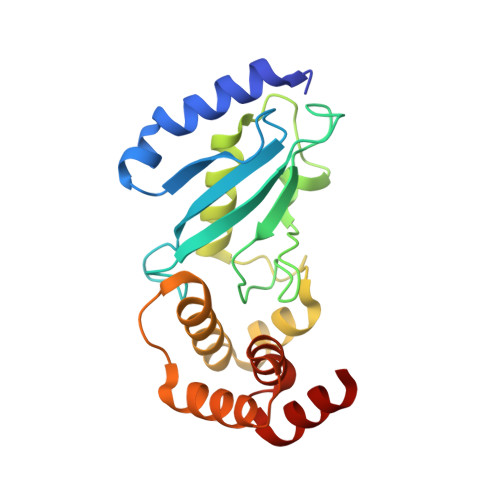 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61086 (Homo sapiens) Explore P61086 Go to UniProtKB: P61086 | |||||
PHAROS: P61086 GTEx: ENSG00000078140 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61086 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquitin | 96 | Homo sapiens | Mutation(s): 0 Gene Names: UBB |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0CG48 (Homo sapiens) Explore P0CG48 Go to UniProtKB: P0CG48 | |||||
PHAROS: P0CG48 GTEx: ENSG00000150991 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0CG48 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 102.48 | α = 90 |
| b = 50.547 | β = 125.01 |
| c = 71.323 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |