Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Laponogov, I., Pan, X.-S., Veselkov, D.A., McAuley, K.E., Fisher, L.M., Sanderson, M.R.(2010) PLoS One 5: e11338-e11338
- PubMed: 20596531
- DOI: https://doi.org/10.1371/journal.pone.0011338
- Primary Citation of Related Structures:
3K9F, 3KSA, 3KSB, 3LTN - PubMed Abstract:
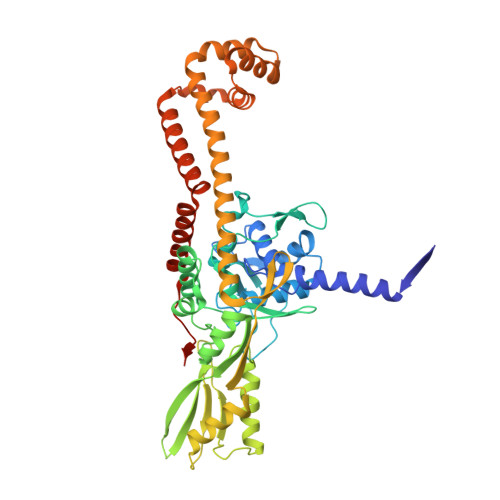
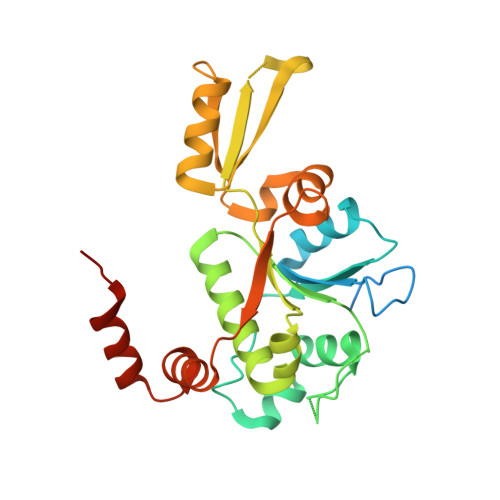
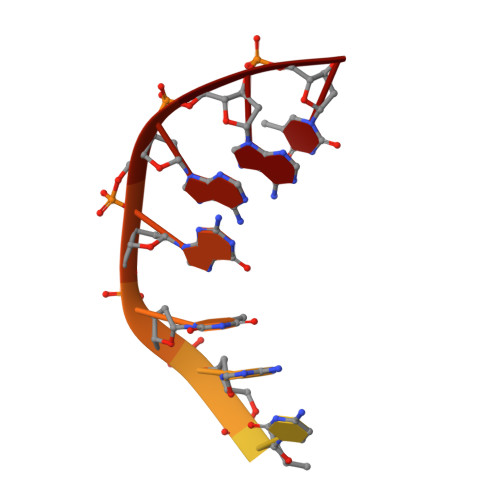
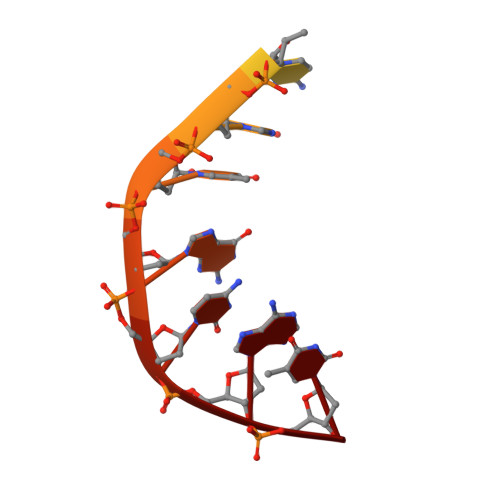
Type II DNA topoisomerases are ubiquitous enzymes with essential functions in DNA replication, recombination and transcription. They change DNA topology by forming a transient covalent cleavage complex with a gate-DNA duplex that allows transport of a second duplex though the gate. Despite its biological importance and targeting by anticancer and antibacterial drugs, cleavage complex formation and reversal is not understood for any type II enzyme. To address the mechanism, we have used X-ray crystallography to study sequential states in the formation and reversal of a DNA cleavage complex by topoisomerase IV from Streptococcus pneumoniae, the bacterial type II enzyme involved in chromosome segregation. A high resolution structure of the complex captured by a novel antibacterial dione reveals two drug molecules intercalated at a cleaved B-form DNA gate and anchored by drug-specific protein contacts. Dione release generated drug-free cleaved and resealed DNA complexes in which the DNA gate instead adopts an unusual A/B-form helical conformation with a Mg(2+) ion repositioned to coordinate each scissile phosphodiester group and promote reversible cleavage by active-site tyrosines. These structures, the first for putative reaction intermediates of a type II topoisomerase, suggest how a type II enzyme reseals DNA during its normal reaction cycle and illuminate aspects of drug arrest important for the development of new topoisomerase-targeting therapeutics.
- Randall Division of Cell and Molecular Biophysics, King's College London, London, United Kingdom.
Organizational Affiliation: