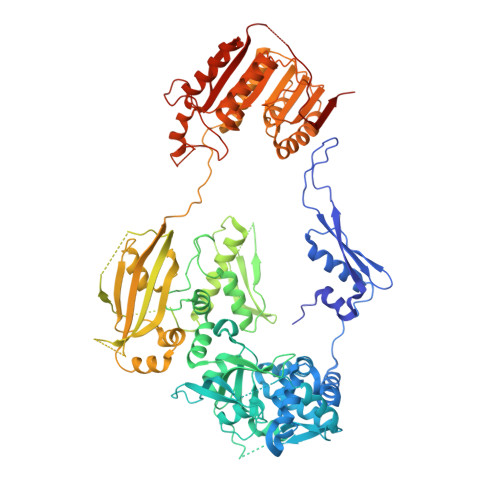
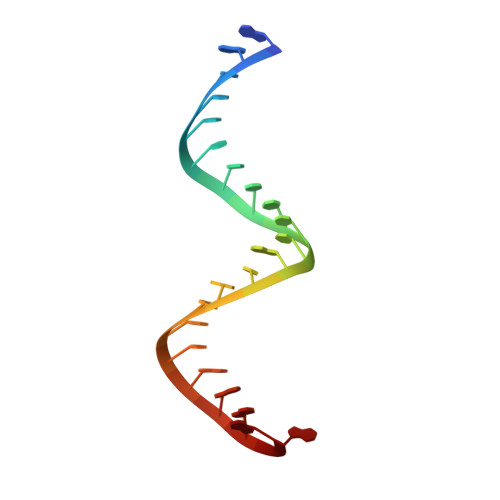
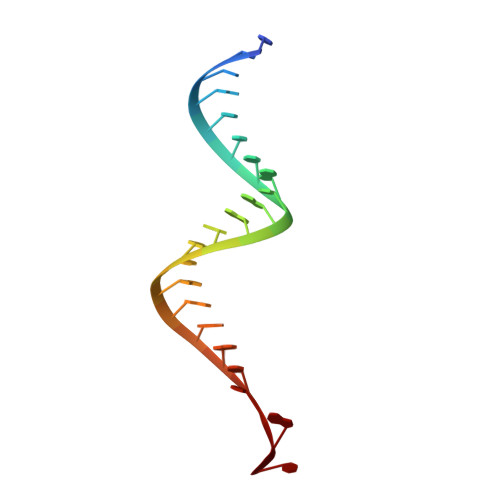
Structural insights into mechanisms of the small RNA methyltransferase HEN1.
Huang, Y., Ji, L., Huang, Q., Vassylyev, D.G., Chen, X., Ma, J.B.(2009) Nature 461: 823-827
- PubMed: 19812675
- DOI: https://doi.org/10.1038/nature08433
- Primary Citation of Related Structures:
3HTX - PubMed Abstract:
RNA silencing is a conserved regulatory mechanism in fungi, plants and animals that regulates gene expression and defence against viruses and transgenes. Small silencing RNAs of approximately 20-30 nucleotides and their associated effector proteins, the Argonaute family proteins, are the central components in RNA silencing. A subset of small RNAs, such as microRNAs and small interfering RNAs (siRNAs) in plants, Piwi-interacting RNAs in animals and siRNAs in Drosophila, requires an additional crucial step for their maturation; that is, 2'-O-methylation on the 3' terminal nucleotide. A conserved S-adenosyl-l-methionine-dependent RNA methyltransferase, HUA ENHANCER 1 (HEN1), and its homologues are responsible for this specific modification. Here we report the 3.1 A crystal structure of full-length HEN1 from Arabidopsis in complex with a 22-nucleotide small RNA duplex and cofactor product S-adenosyl-l-homocysteine. Highly cooperative recognition of the small RNA substrate by multiple RNA binding domains and the methyltransferase domain in HEN1 measures the length of the RNA duplex and determines the substrate specificity. Metal ion coordination by both 2' and 3' hydroxyls on the 3'-terminal nucleotide and four invariant residues in the active site of the methyltransferase domain suggests a novel Mg(2+)-dependent 2'-O-methylation mechanism.
- Department of Biochemistry and Molecular Genetics, Schools of Medicine and Dentistry, University of Alabama at Birmingham, Birmingham, Alabama 35294, USA.
Organizational Affiliation: