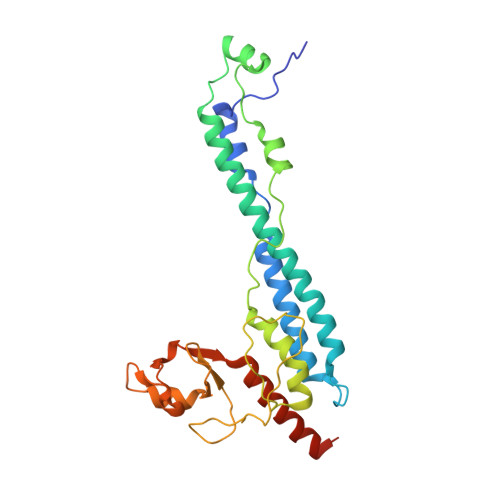
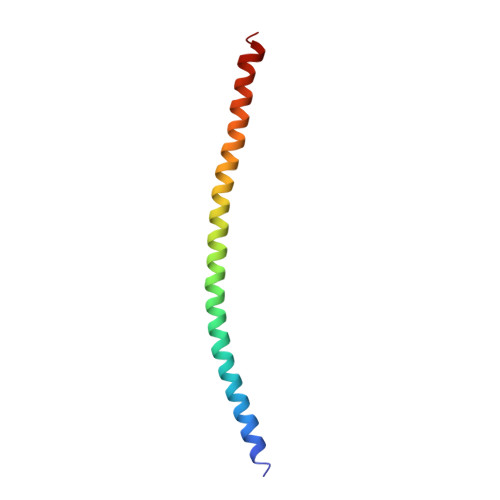Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex.
Duan, X., Sarangi, P., Liu, X., Rangi, G.K., Zhao, X., Ye, H.(2009) Mol Cell 35: 657-668
- PubMed: 19748359
- DOI: https://doi.org/10.1016/j.molcel.2009.06.032
- Primary Citation of Related Structures:
3HTK - PubMed Abstract:
The Smc5/6 complex is an evolutionarily conserved chromosomal ATPase required for cell growth and DNA repair. Its Mms21 subunit supports both functions by docking to the arm region of Smc5 and providing SUMO ligase activity. Here, we report the crystal structure of Mms21 in complex with the Smc5 arm. Our structure revealed two distinct structural and functional domains of the Smc5-bound Mms21: its N-terminal half is dedicated to Smc5 binding by forming a helix bundle with a coiled-coil structure of Smc5; its C-terminal half includes the SUMO ligase domain, which adopts a new type of RING E3 structure. Mutagenesis and structural analyses showed that the Mms21-Smc5 interface is required for cell growth and resistance to DNA damage, while the unique Mms21 RING domain confers specificity to the SUMO E2-E3 interaction. Through structure-based dissection of Mms21 functions, our studies establish a framework for understanding its roles in the Smc5/6 complex.
- James Graham Brown Cancer Center, University of Louisville, KY 40202, USA.
Organizational Affiliation: