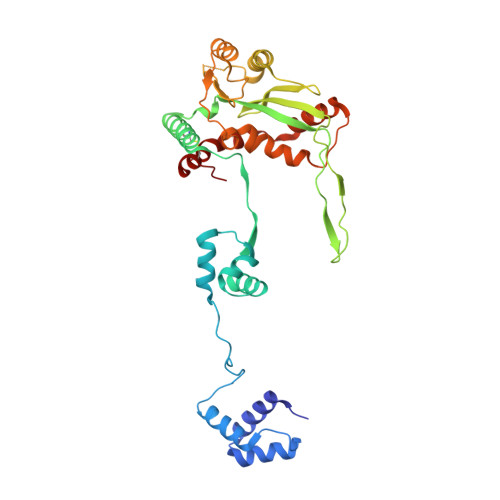
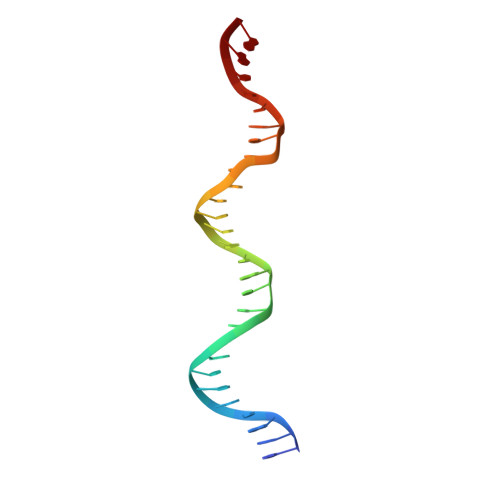
Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote
Richardson, J.M., Colloms, S.D., Finnegan, D.J., Walkinshaw, M.D.(2009) Cell 138: 1096-1108
- PubMed: 19766564
- DOI: https://doi.org/10.1016/j.cell.2009.07.012
- Primary Citation of Related Structures:
3HOS, 3HOT - PubMed Abstract:
A key step in cut-and-paste DNA transposition is the pairing of transposon ends before the element is excised and inserted at a new site in its host genome. Crystallographic analyses of the paired-end complex (PEC) formed from precleaved transposon ends and the transposase of the eukaryotic element Mos1 reveals two parallel ends bound to a dimeric enzyme. The complex has a trans arrangement, with each transposon end recognized by the DNA binding region of one transposase monomer and by the active site of the other monomer. Two additional DNA duplexes in the crystal indicate likely binding sites for flanking DNA. Biochemical data provide support for a model of the target capture complex and identify Arg186 to be critical for target binding. Mixing experiments indicate that a transposase dimer initiates first-strand cleavage and suggest a pathway for PEC formation.
- School of Biological Sciences, University of Edinburgh, Edinburgh, Scotland. jrichard@staffmail.ed.ac.uk
Organizational Affiliation: