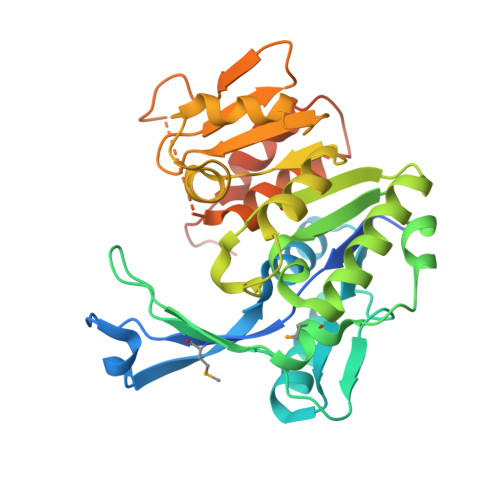
Crystal structure of a fructokinase homolog from Halothermothrix orenii
Chua, T.K., Seetharaman, J., Kasprzak, J.M., Ng, C., Patel, B.K., Love, C., Bujnicki, J.M., Sivaraman, J.(2010) J Struct Biol 171: 397-401
- PubMed: 20493950
- DOI: https://doi.org/10.1016/j.jsb.2010.05.007
- Primary Citation of Related Structures:
3HJ6 - PubMed Abstract:
Fructokinase (FRK; EC 2.7.1.4) catalyzes the phosphorylation of d-fructose to d-fructose 6-phosphate (F6P). This irreversible and near rate-limiting step is a central and regulatory process in plants and bacteria, which channels fructose into a metabolically active state for glycolysis. Towards understanding the mechanism of FRK, here we report the crystal structure of a FRK homolog from a thermohalophilic bacterium Halothermothrixorenii (Hore_18220 in sequence databases). The structure of the Hore_18220 protein reveals a catalytic domain with a Rossmann-like fold and a beta-sheet "lid" for dimerization. Based on comparison of Hore_18220 to structures of related proteins, we propose its mechanism of action, in which the lid serves to regulate access to the substrate binding sites. Close relationship of Hore_18220 and plant FRK enzymes allows us to propose a model for the structure and function of FRKs.
- Department of Biological Sciences, 14 Science Drive 4, National University of Singapore, Singapore 117543, Singapore.
Organizational Affiliation: