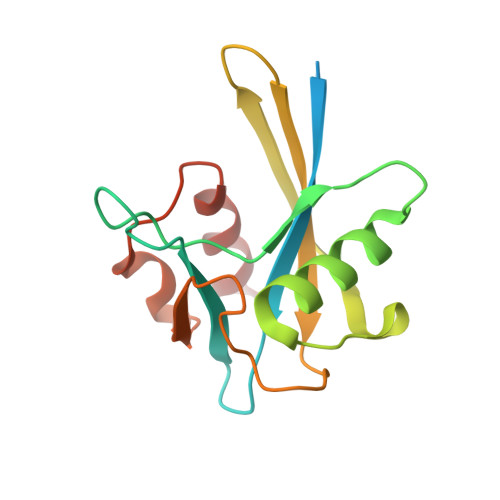
Structure of a Nudix hydrolase (MutT) in the Mg(2+)-bound state from Bartonella henselae, the bacterium responsible for cat scratch fever.
Buchko, G.W., Edwards, T.E., Abendroth, J., Arakaki, T.L., Law, L., Napuli, A.J., Hewitt, S.N., Van Voorhis, W.C., Stewart, L.J., Staker, B.L., Myler, P.J.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1078-1083
- PubMed: 21904053
- DOI: https://doi.org/10.1107/S1744309111011559
- Primary Citation of Related Structures:
3HHJ - PubMed Abstract:
Cat scratch fever (also known as cat scratch disease and bartonellosis) is an infectious disease caused by the proteobacterium Bartonella henselae following a cat scratch. Although the infection usually resolves spontaneously without treatment in healthy adults, bartonellosis may lead to severe complications in young children and immunocompromised patients, and there is new evidence suggesting that B. henselae may be associated with a broader range of clinical symptoms then previously believed. The genome of B. henselae contains genes for two putative Nudix hydrolases, BH02020 and BH01640 (KEGG). Nudix proteins play an important role in regulating the intracellular concentration of nucleotide cofactors and signaling molecules. The amino-acid sequence of BH02020 is similar to that of the prototypical member of the Nudix superfamily, Escherichia coli MutT, a protein that is best known for its ability to neutralize the promutagenic compound 7,8-dihydro-8-oxoguanosine triphosphate. Here, the crystal structure of BH02020 (Bh-MutT) in the Mg(2+)-bound state was determined at 2.1 Å resolution (PDB entry 3hhj). As observed in all Nudix hydrolase structures, the α-helix of the highly conserved `Nudix box' in Bh-MutT is one of two helices that sandwich a four-stranded mixed β-sheet with the central two β-strands parallel to each other. The catalytically essential divalent cation observed in the Bh-MutT structure, Mg(2+), is coordinated to the side chains of Glu57 and Glu61. The structure is not especially robust; a temperature melt obtained using circular dichroism spectroscopy shows that Bh-MutT irreversibly unfolds and precipitates out of solution upon heating, with a T(m) of 333 K.
- Seattle Structural Genomics Center for Infectious Disease, http://www.ssgcid.org, USA. garry.buchko@pnnl.gov
Organizational Affiliation: