Crystal Structure of Ctp Pyrophosphohydrolase from Bacteroides fragilis
Patskovsky, Y., Romero, R., Gilmore, M., Do, J., Wasserman, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative CTP pyrophosphohydrolase | 140 | Bacteroides fragilis NCTC 9343 | Mutation(s): 0 Gene Names: BF2916 EC: 3.6.1.55 | 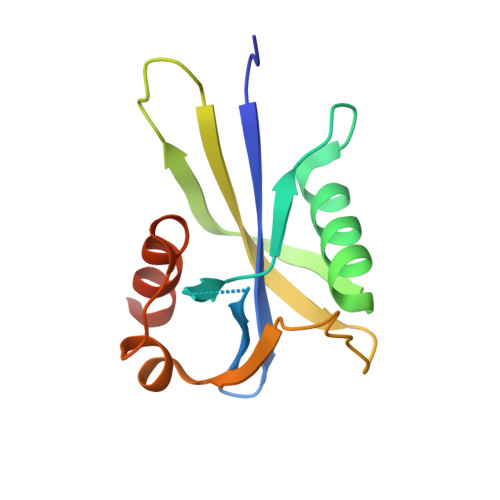 | |
UniProt | |||||
Find proteins for Q5LBB1 (Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / LMG 10263 / NCTC 9343 / Onslow / VPI 2553 / EN-2)) Explore Q5LBB1 Go to UniProtKB: Q5LBB1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5LBB1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.138 | α = 90 |
| b = 62.275 | β = 90 |
| c = 59.777 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345 | data collection |
| SHELXCD | phasing |
| SHELXE | model building |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |