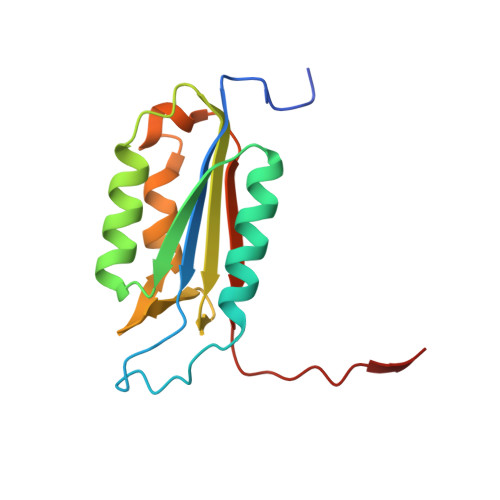
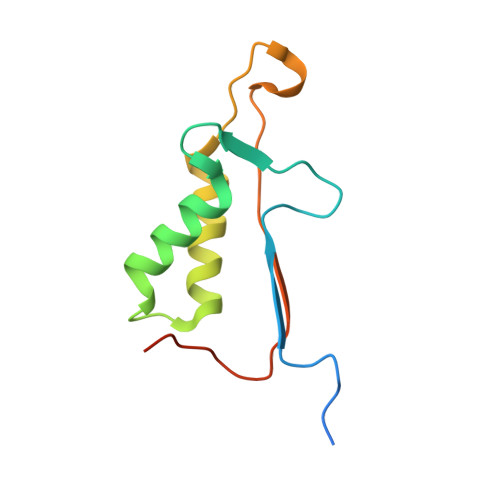
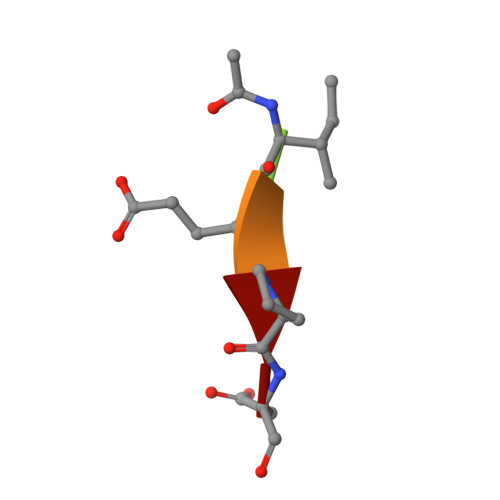
Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Fang, B., Fu, G., Agniswamy, J., Harrison, R.W., Weber, I.T.(2009) Apoptosis 14: 741-752
- PubMed: 19283487
- DOI: https://doi.org/10.1007/s10495-009-0333-y
- Primary Citation of Related Structures:
3GJQ, 3GJR, 3GJS, 3GJT - PubMed Abstract:
Caspase-3 recognition of various P4 residues in its numerous protein substrates was investigated by crystallography, kinetics, and calculations on model complexes. Asp is the most frequent P4 residue in peptide substrates, although a wide variety of P4 residues are found in the cellular proteins cleaved by caspase-3. The binding of peptidic inhibitors with hydrophobic P4 residues, or no P4 residue, is illustrated by crystal structures of caspase-3 complexes with Ac-IEPD-Cho, Ac-WEHD-Cho, Ac-YVAD-Cho, and Boc-D(OMe)-Fmk at resolutions of 1.9-2.6 A. The P4 residues formed favorable hydrophobic interactions in two separate hydrophobic regions of the binding site. The side chains of P4 Ile and Tyr form hydrophobic interactions with caspase-3 residues Trp206 and Trp214 within a non-polar pocket of the S4 subsite, while P4 Trp interacts with Phe250 and Phe252 that can also form the S5 subsite. These interactions of hydrophobic P4 residues are distinct from those for polar P4 Asp, which indicates the adaptability of caspase-3 for binding diverse P4 residues. The predicted trends in peptide binding from molecular models had high correlation with experimental values for peptide inhibitors. Analysis of structural models for the binding of 20 different amino acids at P4 in the aldehyde peptide Ac-XEVD-Cho suggested that the majority of hydrophilic P4 residues interact with Phe250, while hydrophobic residues interact with Trp206, Phe250, and Trp214. Overall, the S4 pocket of caspase-3 exhibits flexible adaptation for different residues and the new structures and models, especially for hydrophobic P4 residues, will be helpful for the design of caspase-3 based drugs.
- Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA.
Organizational Affiliation: