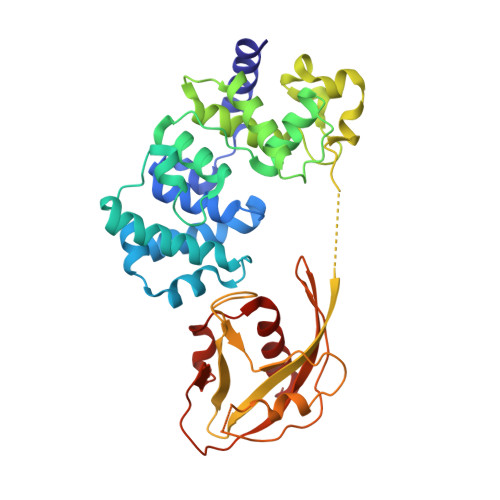
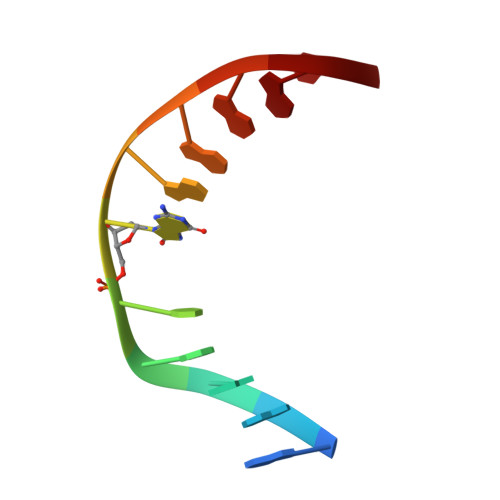
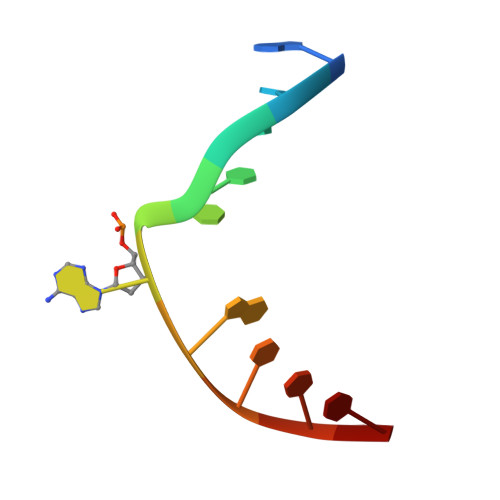
Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase.
Lee, S., Verdine, G.L.(2009) Proc Natl Acad Sci U S A 106: 18497-18502
- PubMed: 19841264
- DOI: https://doi.org/10.1073/pnas.0902908106
- Primary Citation of Related Structures:
3G0Q - PubMed Abstract:
Adenine DNA glycosylase catalyzes the glycolytic removal of adenine from the promutagenic A.oxoG base pair in DNA. The general features of DNA recognition by an adenine DNA glycosylase, Bacillus stearothermophilus MutY, have previously been revealed via the X-ray structure of a catalytically inactive mutant protein bound to an A:oxoG-containing DNA duplex. Although the structure revealed the substrate adenine to be, as expected, extruded from the DNA helix and inserted into an extrahelical active site pocket on the enzyme, the substrate adenine engaged in no direct contacts with active site residues. This feature was paradoxical, because other glycosylases have been observed to engage their substrates primarily through direct contacts. The lack of direct contacts in the case of MutY suggested that either MutY uses a distinctive logic for substrate recognition or that the X-ray structure had captured a noncatalytically competent state in lesion recognition. To gain further insight into this issue, we crystallized wild-type MutY bound to DNA containing a catalytically inactive analog of 2'-deoxyadenosine in which a single 2'-H atom was replaced by fluorine. The structure of this fluorinated lesion-recognition complex (FLRC) reveals the substrate adenine buried more deeply into the active site pocket than in the prior structure and now engaged in multiple direct hydrogen bonding and hydrophobic interactions. This structure appears to capture the catalytically competent state of adenine DNA glycosylases, and it suggests a catalytic mechanism for this class of enzymes, one in which general acid-catalyzed protonation of the nucleobase promotes glycosidic bond cleavage.
- Departments of Stem Cell and Regenerative Biology, Harvard University, 12 Oxford Street, Cambridge, MA 02138, USA.
Organizational Affiliation: