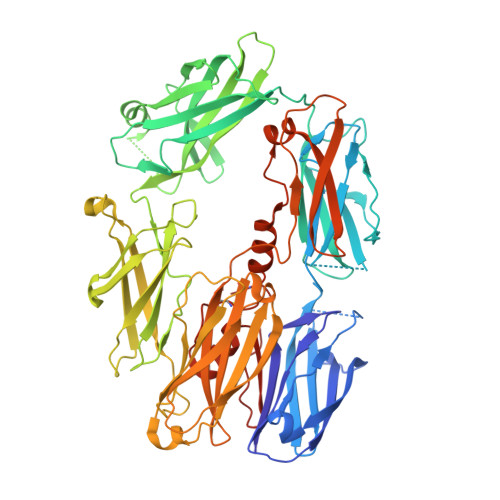
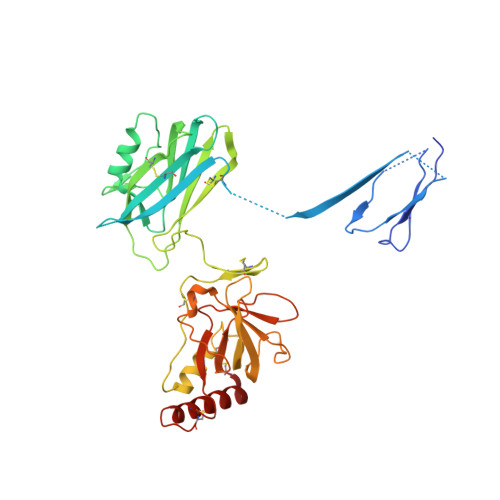
The Crystal Structure of Cobra Venom Factor, a Cofactor for C3- and C5-Convertase CVFBb.
Krishnan, V., Ponnuraj, K., Xu, Y., Macon, K., Volanakis, J.E., Narayana, S.V.(2009) Structure 17: 611-619
- PubMed: 19368894
- DOI: https://doi.org/10.1016/j.str.2009.01.015
- Primary Citation of Related Structures:
3FRP - PubMed Abstract:
Cobra venom factor (CVF) is a functional analog of human complement component C3b, the active fragment of C3. Similar to C3b, in human and mammalian serum, CVF binds factor B, which is then cleaved by factor D, giving rise to the CVFBb complex that targets the same scissile bond in C3 as the authentic complement convertases C4bC2a and C3bBb. Unlike the latter, CVFBb is a stable complex and an efficient C5 convertase. We solved the crystal structure of CVF, isolated from Naja naja kouthia venom, at 2.6 A resolution. The CVF crystal structure, an intermediate between C3b and C3c, lacks the TED domain and has the CUB domain in an identical position to that seen in C3b. The similarly positioned CUB and slightly displaced C345c domains of CVF could play a vital role in the formation of C3 convertases by providing important primary binding sites for factor B.
- University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Organizational Affiliation: