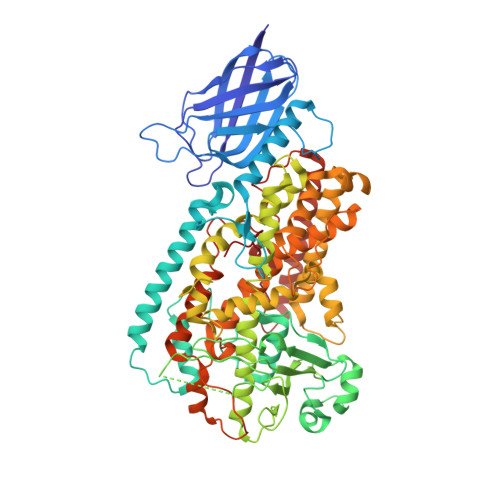
The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Neau, D.B., Gilbert, N.C., Bartlett, S.G., Boeglin, W., Brash, A.R., Newcomer, M.E.(2009) Biochemistry 48: 7906-7915
- PubMed: 19594169
- DOI: https://doi.org/10.1021/bi900084m
- Primary Citation of Related Structures:
3FG1, 3FG3, 3FG4 - PubMed Abstract:
Lipoxygenases (LOX) play pivotal roles in the biosynthesis of leukotrienes and other biologically active eicosanoids derived from arachidonic acid. A mechanistic understanding of substrate recognition, when lipoxygenases that recognize the same substrate generate different products, can be used to help guide the design of enzyme-specific inhibitors. We report here the 1.85 A resolution structure of an 8R-lipoxygenase from Plexaura homomalla, an enzyme with a sequence approximately 40% identical to that of human 5-LOX. The structure reveals a U-shaped channel, defined by invariant amino acids, that would allow substrate access to the catalytic iron. We demonstrate that mutations within the channel significantly impact enzyme activity and propose a novel model for substrate binding potentially applicable to other members of this enzyme family.
- Department of Biological Sciences, Louisiana State University, Baton Rouge, Louisiana 70803, USA.
Organizational Affiliation: