Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Liu, B., Chen, Y., Doukov, T., Soltis, S.M., Stout, C.D., Fee, J.A.(2009) Biochemistry 48: 820-826
- PubMed: 19140675
- DOI: https://doi.org/10.1021/bi801759a
- Primary Citation of Related Structures:
3EH3, 3EH4, 3EH5 - PubMed Abstract:
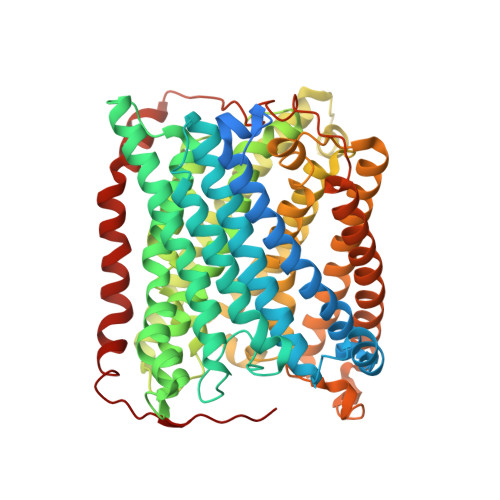
Three paths for obtaining crystals of reduced (II-E4Q/I-K258R) cytochrome ba(3) are described, and the structures of these are reported at approximately 2.8-3.0 A resolution. Microspectrophotometry of single crystals of Thermus ba(3) oxidase at 100 K was used to show that crystals of the oxidized enzyme are reduced in an intense X-ray (beam line 7-1 at the Stanford Synchrotron Radiation Laboratory), being nearly complete in 1 min. The previously reported structures of ba(3) (Protein Data Bank entries 1EHK and 1XME ), having a crystallographically detectable water between the Cu(B) and Fe(a3) metals of the dinuclear center, actually represent the X-ray radiation-reduced enzyme. Dithionite-reduced crystals or crystals formed from dithionite-reduced enzyme revealed the absence of the above-mentioned water and an increase in the Cu(B)-Fe(a3) distance of approximately 0.3 A. The new structures are discussed in terms of enzyme function. An unexpected optical absorption envelope at approximately 590 nm is also reported. This spectral feature is tentatively thought to arise from a five-coordinate, low-spin, ferrous heme a(3) that is trapped in the frozen crystals.
- Department of Molecular Biology, The Scripps Research Institute, MB-8, 10550 North Torrey Pines Road, La Jolla, California 92037, USA.
Organizational Affiliation: