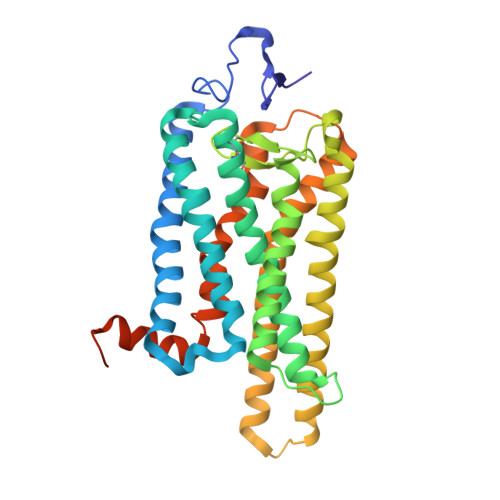
Crystal structure of the ligand-free G-protein-coupled receptor opsin
Park, J.H., Scheerer, P., Hofmann, K.P., Choe, H.-W., Ernst, O.P.(2008) Nature 454: 183-187
- PubMed: 18563085
- DOI: https://doi.org/10.1038/nature07063
- Primary Citation of Related Structures:
3CAP - PubMed Abstract:
In the G-protein-coupled receptor (GPCR) rhodopsin, the inactivating ligand 11-cis-retinal is bound in the seven-transmembrane helix (TM) bundle and is cis/trans isomerized by light to form active metarhodopsin II. With metarhodopsin II decay, all-trans-retinal is released, and opsin is reloaded with new 11-cis-retinal. Here we present the crystal structure of ligand-free native opsin from bovine retinal rod cells at 2.9 ångström (A) resolution. Compared to rhodopsin, opsin shows prominent structural changes in the conserved E(D)RY and NPxxY(x)(5,6)F regions and in TM5-TM7. At the cytoplasmic side, TM6 is tilted outwards by 6-7 A, whereas the helix structure of TM5 is more elongated and close to TM6. These structural changes, some of which were attributed to an active GPCR state, reorganize the empty retinal-binding pocket to disclose two openings that may serve the entry and exit of retinal. The opsin structure sheds new light on ligand binding to GPCRs and on GPCR activation.
- Institut für Medizinische Physik und Biophysik (CC2), Charité-Universitätsmedizin Berlin, Charitéplatz 1, D-10117 Berlin, Germany.
Organizational Affiliation: