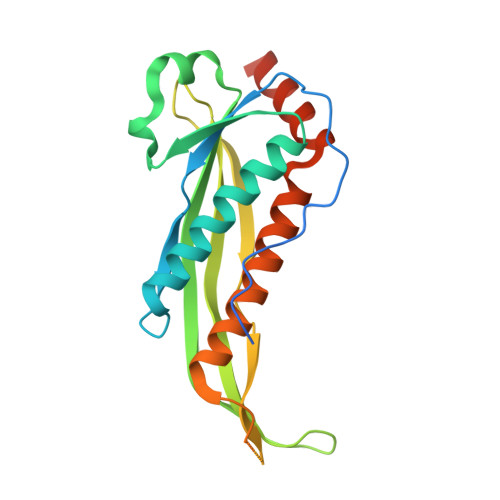
Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori.
Bonanno, J.B., Dickey, J., Bain, K.T., McKenzie, C., Romero, R., Smith, D., Wasserman, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.