A molecular mechanism for copper transportation to tyrosinase that is assisted by a metallochaperone, caddie protein
Matoba, Y., Bando, N., Oda, K., Noda, M., Higashikawa, F., Kumagai, T., Sugiyama, M.(2011) J Biological Chem 286: 30219-30231
- PubMed: 21730070
- DOI: https://doi.org/10.1074/jbc.M111.256818
- Primary Citation of Related Structures:
3AWS, 3AWT, 3AWU, 3AWV, 3AWW, 3AWX, 3AWY, 3AWZ, 3AX0 - PubMed Abstract:
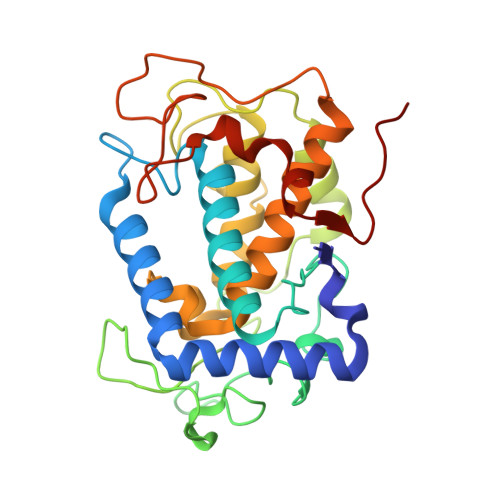
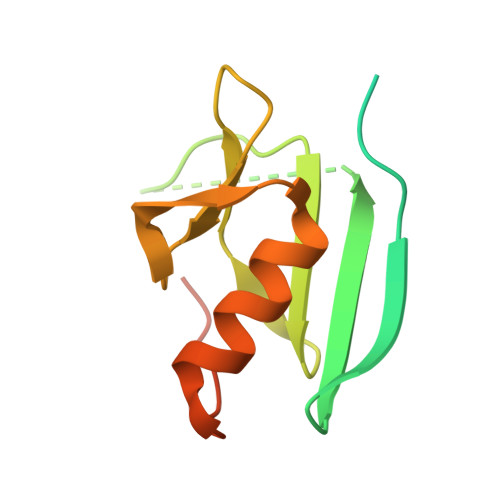
The Cu(II)-soaked crystal structure of tyrosinase that is present in a complex with a protein, designated "caddie," which we previously determined, possesses two copper ions at its catalytic center. We had identified two copper-binding sites in the caddie protein and speculated that copper bound to caddie may be transported to the tyrosinase catalytic center. In our present study, at a 1.16-1.58 Å resolution, we determined the crystal structures of tyrosinase complexed with caddie prepared by altering the soaking time of the copper ion and the structures of tyrosinase complexed with different caddie mutants that display little or no capacity to activate tyrosinase. Based on these structures, we propose a molecular mechanism by which two copper ions are transported to the tyrosinase catalytic center with the assistance of caddie acting as a metallochaperone.
- Department of Molecular Microbiology and Biotechnology, Graduate School of Biomedical Sciences, Hiroshima University, Minami-ku, Hiroshima, Japan.
Organizational Affiliation: