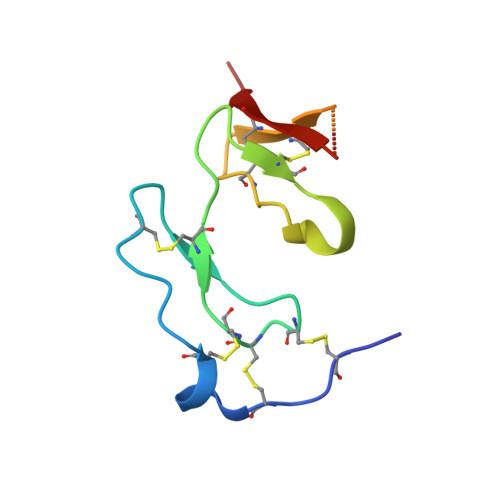
Structural and Biochemical Characterization of Native and Recombinant Single Insulin-Like Growth Factor-Binding Domain Protein (Sibd-1) from the Central American Hunting Spider Cupiennius Salei (Ctenidae).
Trachsel, C., Widmer, C., Kampfer, U., Buhr, C., Baumann, T., Kuhn-Nentwig, L., Schurch, S., Schaller, J., Baumann, U.(2012) Proteins 80: 2323
- PubMed: 22622866
- DOI: https://doi.org/10.1002/prot.24119
- Primary Citation of Related Structures:
3ZXB, 3ZXC - PubMed Abstract:
Cupiennius salei single insulin-like growth factor binding domain protein (SIBD-1) is an 8.6 kDa Cys-, Pro-, and Gly-rich protein, discovered in the hemocytes of the Central American hunting spider Cupiennius salei. SIBD-1 exhibits high sequence similarity to the N-terminal domain of the insulin-like growth factor-binding protein superfamily and has been reported to play an important role in the spider's immune system. Here, the recombinant expression and the elucidation of the three-dimensional structure of recombinant SIBD-1 and the characterization of the sugar moiety at Thr2 of native SIBD-1 is described in detail.
- Department of Chemistry and Biochemistry, University of Bern, Bern, Switzerland.
Organizational Affiliation: