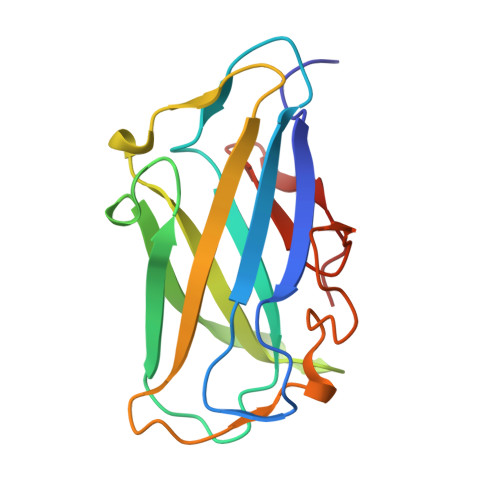
Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Yaniv, O., Halfon, Y., Shimon, L.J.W., Bayer, E.A., Lamed, R., Frolow, F.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 8
- PubMed: 22232162
- DOI: https://doi.org/10.1107/S174430911104807X
- Primary Citation of Related Structures:
3ZQW, 3ZU8, 3ZUC - PubMed Abstract:
The carbohydrate-binding module (CBM) of the major scaffoldin subunit ScaA of the cellulosome of Acetivibrio cellulolyticus is classified as a family 3b CBM and binds strongly to cellulose. The CBM3b was overexpressed, purified and crystallized, and its three-dimensional structure was determined. The structure contained a nickel-binding site located at the N-terminal region in addition to a 'classical' CBM3b calcium-binding site. The structure was also determined independently by the SAD method using data collected at the Ni-absorption wavelength of 1.48395 Å and even at a wavelength of 0.97625 Å in a favourable case. The new scaffoldin-borne CBM3 structure reported here provides clear evidence for the proposition that a family 3b CBM may be accommodated in scaffoldin subunits and functions as the major substrate-binding entity of the cellulosome assembly.
- Department of Molecular Microbiology and Biotechnology, Tel Aviv University, 69978 Tel Aviv, Israel.
Organizational Affiliation: