Selective Aurora Kinase Inhibitors Identified Using a Taxol- Induced Checkpoint Sensitivity Screen.
Kwiatkowski, N., Deng, X., Wang, J., Tan, L., Villa, F., Santaguida, S., Huang, H.C., Mitchison, T., Musacchio, A., Gray, N.(2012) ACS Chem Biol 7: 185
- PubMed: 21992004
- DOI: https://doi.org/10.1021/cb200305u
- Primary Citation of Related Structures:
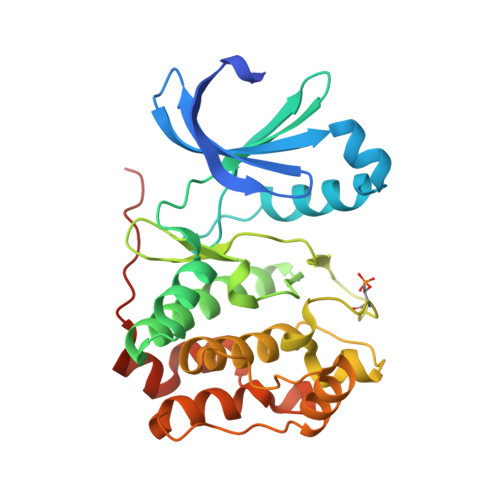
3ZTX - PubMed Abstract:
The members of the Aurora kinase family play critical roles in the regulation of the cell cycle and mitotic spindle assembly and have been intensively investigated as potential targets for a new class of anticancer drugs. We describe a new highly potent and selective class of Aurora kinase inhibitors discovered using a phenotypic cellular screen. Optimized inhibitors display many of the hallmarks of Aurora inhibition including endoreduplication, polyploidy, and loss of cell viability in cancer cells. Structure-activity relationships with respect to kinome-wide selectivity and guided by an Aurora B co-crystal structure resulted in the identification of key selectivity determinants and discovery of a subseries with selectivity toward Aurora A. A direct comparison of biochemical and cellular profiles with respect to published Aurora inhibitors including VX-680, AZD1152, MLN8054, and a pyrimidine-based compound from Genentech demonstrates that compounds 1 and 3 will become valuable additional pharmacological probes of Aurora-dependent functions.
- Department of Cancer Biology, Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts 02115, United States.
Organizational Affiliation: