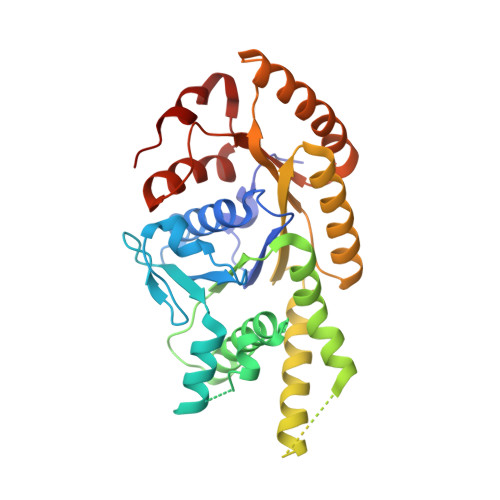
The Mechanism of Membrane-Associated Steps in Tail-Anchored Protein Insertion.
Mariappan, M., Mateja, A., Dobosz, M., Bove, E., Hegde, R.S., Keenan, R.J.(2011) Nature 477: 61
- PubMed: 21866104
- DOI: https://doi.org/10.1038/nature10362
- Primary Citation of Related Structures:
3ZS8, 3ZS9 - PubMed Abstract:
Tail-anchored (TA) membrane proteins destined for the endoplasmic reticulum are chaperoned by cytosolic targeting factors that deliver them to a membrane receptor for insertion. Although a basic framework for TA protein recognition is now emerging, the decisive targeting and membrane insertion steps are not understood. Here we reconstitute the TA protein insertion cycle with purified components, present crystal structures of key complexes between these components and perform mutational analyses based on the structures. We show that a committed targeting complex, formed by a TA protein bound to the chaperone ATPase Get3, is initially recruited to the membrane through an interaction with Get2. Once the targeting complex has been recruited, Get1 interacts with Get3 to drive TA protein release in an ATPase-dependent reaction. After releasing its TA protein cargo, the now-vacant Get3 recycles back to the cytosol concomitant with ATP binding. This work provides a detailed structural and mechanistic framework for the minimal TA protein insertion cycle.
- Cell Biology and Metabolism Program, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: