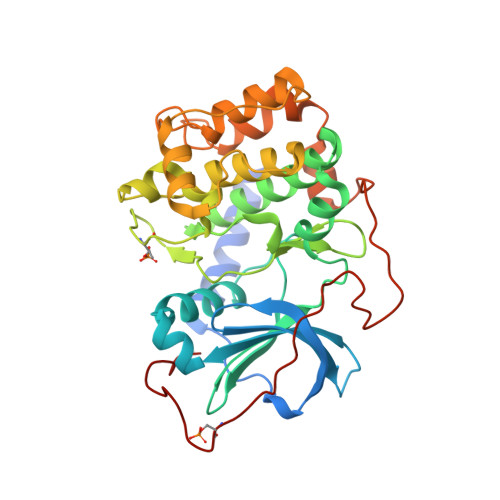
Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Allen, C.E., Chow, C.L., Caldwell, J.J., Westwood, I.M., van Montfort, R.L., Collins, I.(2013) Bioorg Med Chem 21: 5707-5724
- PubMed: 23920481
- DOI: https://doi.org/10.1016/j.bmc.2013.07.021
- Primary Citation of Related Structures:
3ZO1, 3ZO2, 3ZO3, 3ZO4 - PubMed Abstract:
With the success of protein kinase inhibitors as drugs to target cancer, there is a continued need for new kinase inhibitor scaffolds. We have investigated the synthesis and kinase inhibition of new heteroaryl-substituted diazaspirocyclic compounds that mimic ATP. Versatile syntheses of substituted diazaspirocycles through ring-closing metathesis were demonstrated. Diazaspirocycles directly linked to heteroaromatic hinge binder groups provided ligand efficient inhibitors of multiple kinases, suitable as starting points for further optimization. The binding modes of representative diazaspirocyclic motifs were confirmed by protein crystallography. Selectivity profiles were influenced by the hinge binder group and the interactions of basic nitrogen atoms in the scaffold with acidic side-chains of residues in the ATP pocket. The introduction of more complex substitution to the diazaspirocycles increased potency and varied the selectivity profiles of these initial hits through engagement of the P-loop and changes to the spirocycle conformation, demonstrating the potential of these core scaffolds for future application to kinase inhibitor discovery.
- Cancer Research UK Cancer Therapeutics Unit, The Institute of Cancer Research, Sutton, UK.
Organizational Affiliation: