The Crystal Structure of the Lumenal Domain of Erv41P, a Protein Involved in Transport between the Endoplasmic Reticulum and Golgi Apparatus
Biterova, E.I., Svard, M., Possner, D.D.D., Guy, J.E.(2013) J Mol Biol 425: 2208
- PubMed: 23524136
- DOI: https://doi.org/10.1016/j.jmb.2013.03.024
- Primary Citation of Related Structures:
3ZLC - PubMed Abstract:
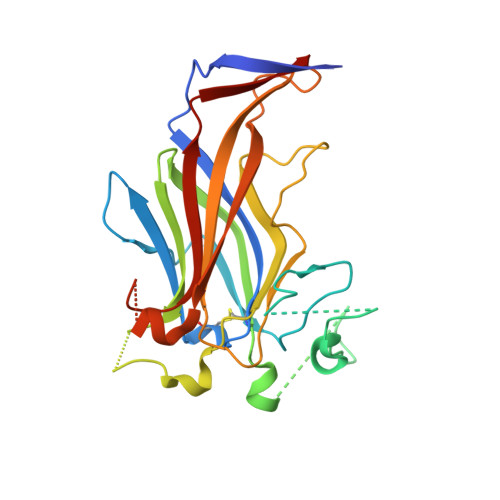
Erv41p is a conserved integral membrane protein that is known to play a role in transport between the endoplasmic reticulum and Golgi apparatus, part of the early secretory pathway of eukaryotes. However, the exact function of the protein is not known, and it shares very low sequence identity with proteins of known structure or function. Here we present the structure of the full lumenal domain of Erv41p from Saccharomyces cerevisiae, determined by X-ray crystallography to a resolution of 2.0Å. The structure reveals the protein to be composed predominantly of two large β-sheets that form a twisted β-sandwich. Comparison to structures in the Protein Data Bank shows that the Erv41p lumenal domain displays only limited similarity to β-sandwich domains of other proteins. Analysis of the surface properties of the protein identifies an extensive patch of negative electrostatic potential on the exposed surface of one of the β-sheets, which likely forms a binding site for a ligand or interaction partner. A predominantly hydrophobic region close to the membrane interface is identified as a likely site for protein-protein interaction. This structure, the first of Erv41p or any of its homologues, provides a new starting point for studies of the roles of Erv41p and its interaction partners in the eukaryotic secretory pathway.
Organizational Affiliation:
Division of Molecular Structural Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Tomtebodavägen 6, 4tr, Stockholm S-171 77, Sweden.