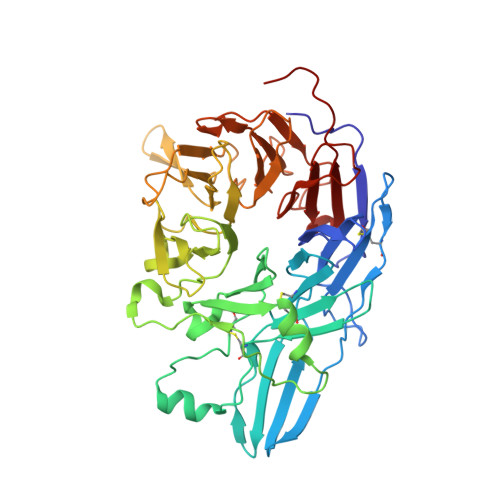
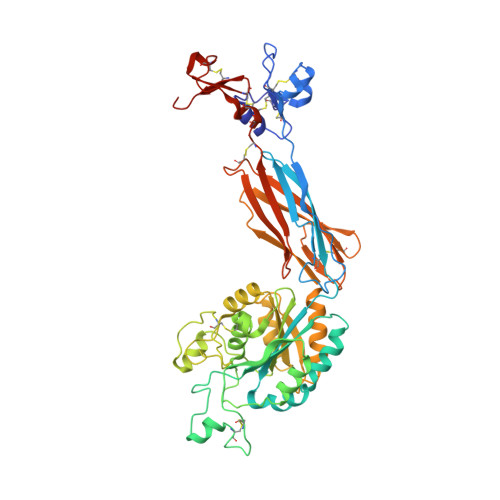
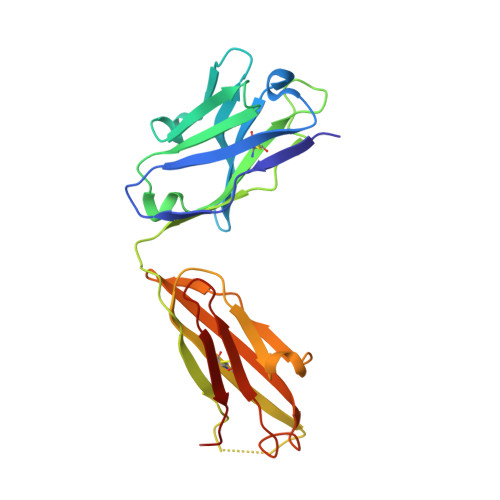
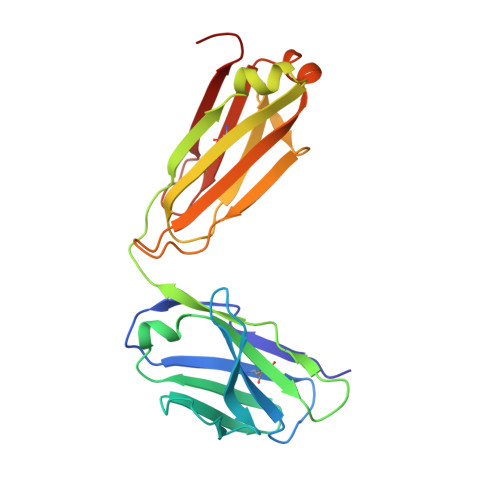
Complete Integrin Headpiece Opening in Eight Steps.
Zhu, J., Zhu, J., Springer, T.A.(2013) J Cell Biol 201: 1053
- PubMed: 23798730
- DOI: https://doi.org/10.1083/jcb.201212037
- Primary Citation of Related Structures:
3ZDX, 3ZDY, 3ZDZ, 3ZE0, 3ZE1, 3ZE2 - PubMed Abstract:
Carefully soaking crystals with Arg-Gly-Asp (RGD) peptides, we captured eight distinct RGD-bound conformations of the αIIbβ3 integrin headpiece. Starting from the closed βI domain conformation, we saw six intermediate βI conformations and finally the fully open βI with the hybrid domain swung out in the crystal lattice. The β1-α1 backbone that hydrogen bonds to the Asp side chain of RGD was the first element to move followed by adjacent to metal ion-dependent adhesion site Ca(2+), α1 helix, α1' helix, β6-α7 loop, α7 helix, and hybrid domain. We define in atomic detail how conformational change was transmitted over long distances in integrins, 40 Å from the ligand binding site to the opposite end of the βI domain and 80 Å to the far end of the hybrid domain. During these movements, RGD slid in its binding groove toward αIIb, and its Arg side chain became ordered. RGD concentration requirements in soaking suggested a >200-fold higher affinity after opening. The thermodynamic cycle shows how higher affinity pays the energetic cost of opening.
- Department of Biological Chemistry and Molecular Pharmacology, Program in Cellular and Molecular Medicine, Children's Hospital Boston, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: