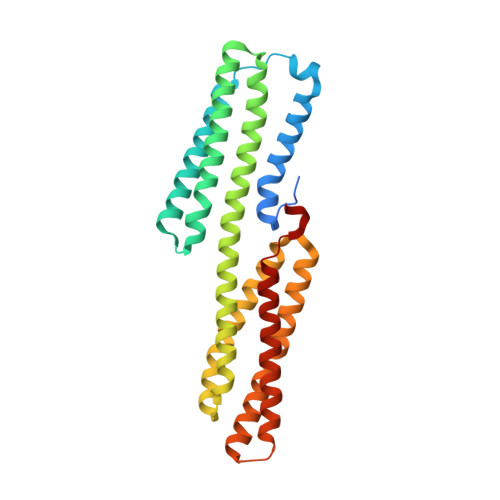
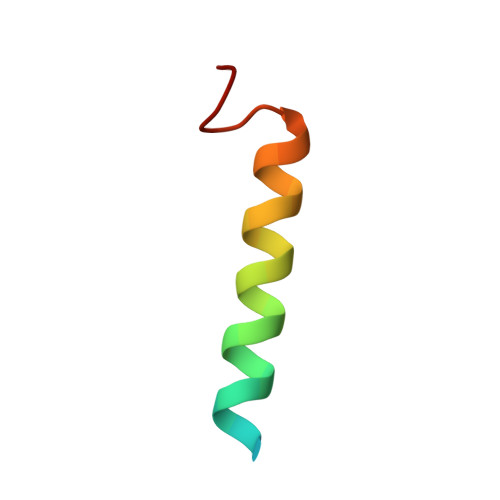
Riam and Vinculin Binding to Talin are Mutually Exclusive and Regulate Adhesion Assembly and Turnover.
Goult, B.T., Zacharchenko, T., Bate, N., Tsang, R., Hey, F., Gingras, A.R., Elliott, P.R., Roberts, G.C., Ballestrem, C., Critchley, D.R., Barsukov, I.L.(2013) J Biological Chem 288: 8238
- PubMed: 23389036
- DOI: https://doi.org/10.1074/jbc.M112.438119
- Primary Citation of Related Structures:
2L10, 2L7A, 2L7N, 2LQG, 3ZDL - PubMed Abstract:
Talin activates integrins, couples them to F-actin, and recruits vinculin to focal adhesions (FAs). Here, we report the structural characterization of the talin rod: 13 helical bundles (R1-R13) organized into a compact cluster of four-helix bundles (R2-R4) within a linear chain of five-helix bundles. Nine of the bundles contain vinculin-binding sites (VBS); R2R3 are atypical, with each containing two VBS. Talin R2R3 also binds synergistically to RIAM, a Rap1 effector involved in integrin activation. Biochemical and structural data show that vinculin and RIAM binding to R2R3 is mutually exclusive. Moreover, vinculin binding requires domain unfolding, whereas RIAM binds the folded R2R3 double domain. In cells, RIAM is enriched in nascent adhesions at the leading edge whereas vinculin is enriched in FAs. We propose a model in which RIAM binding to R2R3 initially recruits talin to membranes where it activates integrins. As talin engages F-actin, force exerted on R2R3 disrupts RIAM binding and exposes the VBS, which recruit vinculin to stabilize the complex.
- Department of Biochemistry, University of Leicester, Lancaster Road, Leicester LE1 9HN, United Kingdom.
Organizational Affiliation: