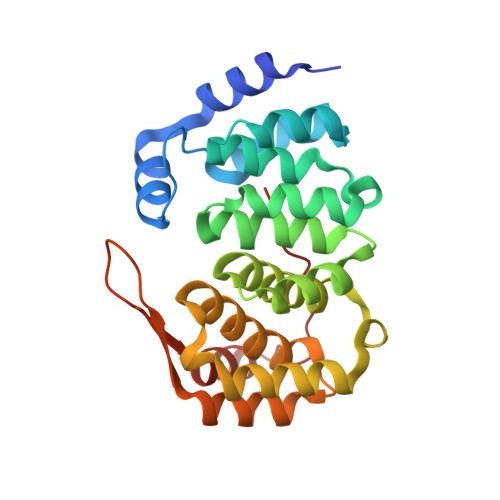
A New Family of Proteins Related to the Heat-Like Repeat DNA Glycosylases with Affinity for Branched DNA Structures.
Backe, P.H., Simm, R., Laerdahl, J.K., Dalhus, B., Fagerlund, A., Okstad, O.A., Rognes, T., Alseth, I., Kolsto, A.-B., Bjoras, M.(2013) J Struct Biol 183: 66
- PubMed: 23623903
- DOI: https://doi.org/10.1016/j.jsb.2013.04.007
- Primary Citation of Related Structures:
3ZBO - PubMed Abstract:
The recently discovered HEAT-like repeat (HLR) DNA glycosylase superfamily is widely distributed in all domains of life. The present bioinformatics and phylogenetic analysis shows that HLR DNA glycosylase superfamily members in the genus Bacillus form three subfamilies: AlkC, AlkD and AlkF/AlkG. The crystal structure of AlkF shows structural similarity with the DNA glycosylases AlkC and AlkD, however neither AlkF nor AlkG display any DNA glycosylase activity. Instead, both proteins have affinity to branched DNA structures such as three-way and Holliday junctions. A unique β-hairpin in the AlkF/AlkG subfamily is most likely inserted into the DNA major groove, and could be a structural determinant regulating DNA substrate affinity. We conclude that AlkF and AlkG represent a new family of HLR proteins with affinity for branched DNA structures.
- Department of Microbiology, Oslo University Hospital and University of Oslo, P.O. Box 4950 Nydalen, 0424 Oslo, Norway.
Organizational Affiliation: