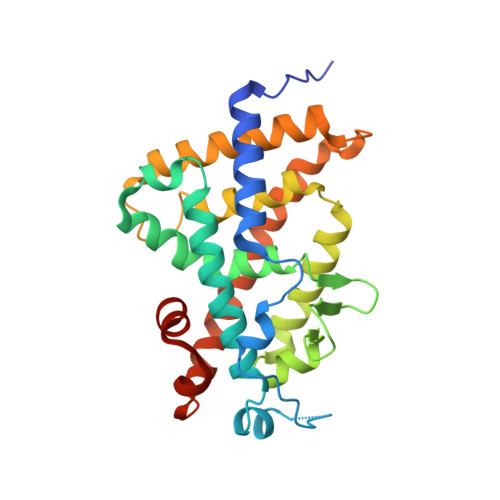
Revisiting the 7,8-cis-vitamin D3 derivatives: synthesis, evaluating the biological activity, and study of the binding configuration
Sawada, D., Saito, H., Kakuda, S., Takimoto-Kamimura, M., Matsumoto, Y., Takagi, K., Ochiai, E., Kittaka, A.To be published.