Purification and preliminary structural studies of hemoglobin from high oxygen affinity species Emu (Dromaius novaehollandiae) at neutral pH
Abubakkar, M.M., Saraboji, K., Ponnuswamy, M.N.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemoglobin subunit alpha-A | 142 | Dromaius novaehollandiae | Mutation(s): 0 | 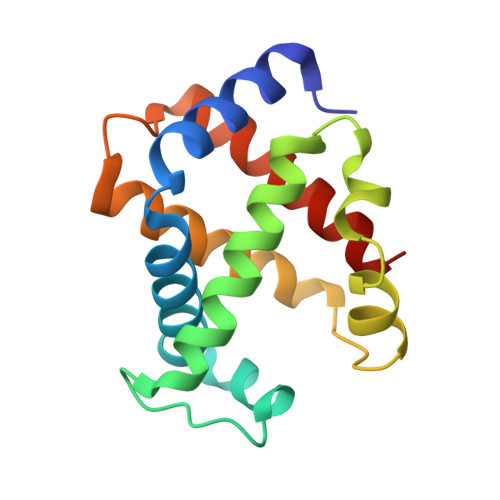 | |
UniProt | |||||
Find proteins for C6L8R0 (Dromaius novaehollandiae) Explore C6L8R0 Go to UniProtKB: C6L8R0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C6L8R0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemoglobin | 146 | Dromaius novaehollandiae | Mutation(s): 0 | 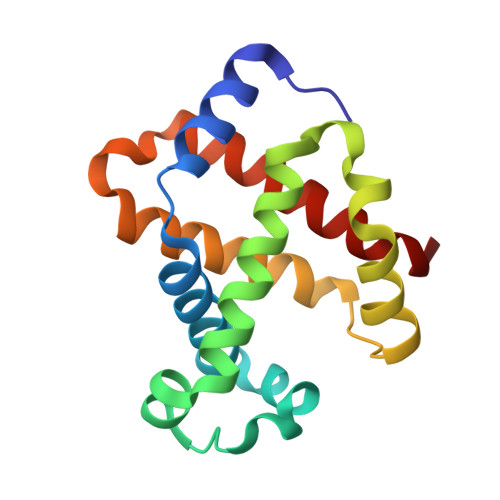 | |
UniProt | |||||
Find proteins for A0A075B5G0 (Dromaius novaehollandiae) Explore A0A075B5G0 Go to UniProtKB: A0A075B5G0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A075B5G0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | E [auth A], G [auth B], I [auth C], K [auth D] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| OXY Query on OXY | F [auth A], H [auth B], J [auth C], L [auth D] | OXYGEN MOLECULE O2 MYMOFIZGZYHOMD-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 66.271 | α = 90 |
| b = 80.01 | β = 90 |
| c = 103.559 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| AUTOMAR | data reduction |
| SCALEPACK | data scaling |