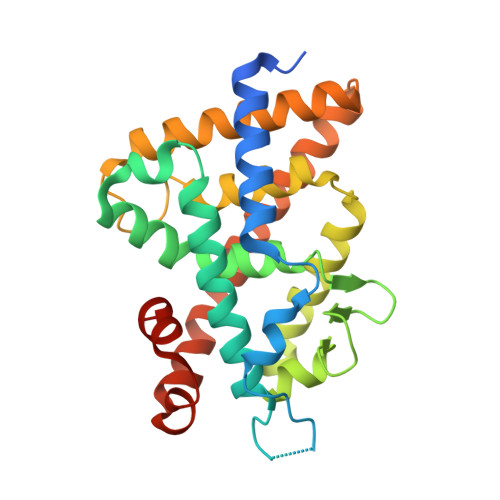
A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
Anami, Y., Itoh, T., Egawa, D., Yoshimoto, N., Yamamoto, K.(2014) J Med Chem 57: 4351-4367
- PubMed: 24742174
- DOI: https://doi.org/10.1021/jm500392t
- Primary Citation of Related Structures:
3WT5, 3WT6, 3WT7 - PubMed Abstract:
We are continuing to study the structural basis of vitamin D receptor (VDR) agonism and antagonism by using 22S-alkyl vitamin D analogues. Here we report the synthesis and biological evaluation of 22R-alkyl analogues and the X-ray crystallographic analysis of vitamin D receptor ligand-binding domain (VDR-LBD) complexed with a 22R-analogue. VDR-LBD complexed with the partial agonist 8a showed that 8a binds to VDR-LBD with two conformations, one of which is the antagonist/VDR-LBD complex structure and the other is the agonist/VDR-LBD complex structure. The results indicate that the partial agonist activity of 8a depends on the sum of antagonistic and agonistic activities caused by the antagonist and agonist binding conformers, respectively. The structural basis observed here must be applicable to the partial agonism of other ligand-dependent nuclear receptors. This is the first report describing the trapping of a conformational subset of the ligand and the nuclear receptor in a single crystal.
- Laboratory of Drug Design and Medicinal Chemistry, Showa Pharmaceutical University , 3-3165 Higashi-Tamagawagakuen, Machida, Tokyo 194-8543, Japan.
Organizational Affiliation: