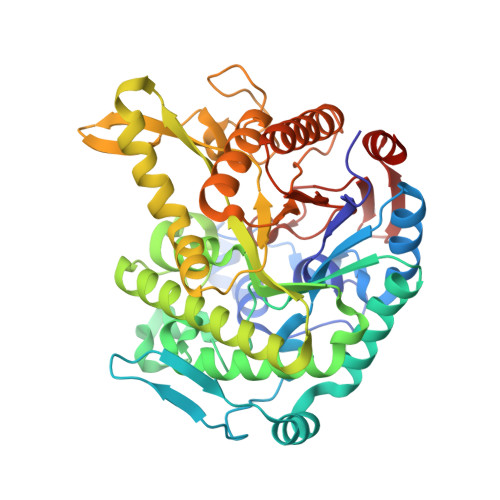
Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Nakabayashi, M., Kataoka, M., Watanabe, M., Ishikawa, K.(2014) Acta Crystallogr F Struct Biol Commun 70: 854-859
- PubMed: 25005077
- DOI: https://doi.org/10.1107/S2053230X14010188
- Primary Citation of Related Structures:
3WQ8 - PubMed Abstract:
One of the β-glucosidases from Pyrococcus furiosus (BGLPf) is found to be a hyperthermophilic tetrameric enzyme that can degrade cellooligosaccharides. Recently, the crystal structures of the tetrameric and dimeric forms were solved. Here, a new monomeric form of BGLPf was constructed by removing the C-terminal region of the enzyme and its crystal structure was solved at a resolution of 2.8 Å in space group P1. It was discovered that the mutant enzyme forms a unique dodecameric structure consisting of two hexameric rings in the asymmetric unit of the crystal. Under biological conditions, the mutant enzyme forms a monomer. This result helps explain how BGLPf has attained its oligomeric structure and thermostability.
- Biomass Refinery Research Center, National Institute of Advanced Industrial Science, 3-11-32 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-0046, Japan.
Organizational Affiliation: