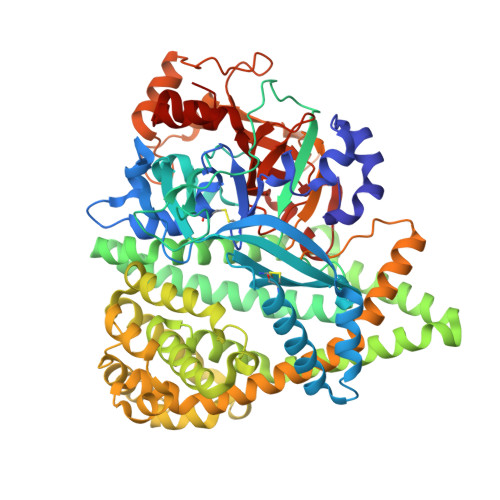
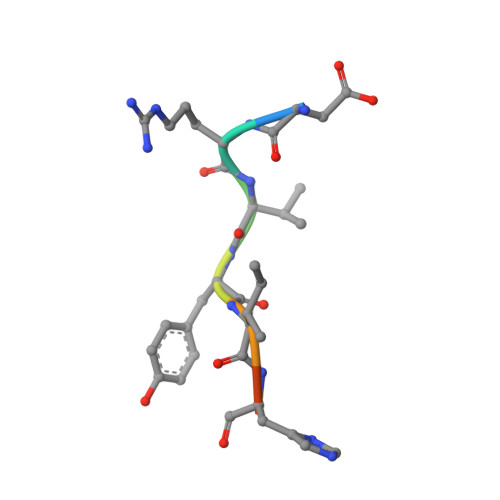
S46 peptidases are the first exopeptidases to be members of clan PA
Sakamoto, Y., Suzuki, Y., Iizuka, I., Tateoka, C., Roppongi, S., Fujimoto, M., Inaka, K., Tanaka, H., Masaki, M., Ohta, K., Okada, H., Nonaka, T., Morikawa, Y., Nakamura, K.T., Ogasawara, W., Tanaka, N.(2014) Sci Rep 4: 4977-4977
- PubMed: 24827749
- DOI: https://doi.org/10.1038/srep04977
- Primary Citation of Related Structures:
3WOI, 3WOJ, 3WOK, 3WOL, 3WOM, 3WON, 3WOO, 3WOP, 3WOQ, 3WOR - PubMed Abstract:
The dipeptidyl aminopeptidase BII (DAP BII) belongs to a serine peptidase family, S46. The amino acid sequence of the catalytic unit of DAP BII exhibits significant similarity to those of clan PA endopeptidases, such as chymotrypsin. However, the molecular mechanism of the exopeptidase activity of family S46 peptidase is unknown. Here, we report crystal structures of DAP BII. DAP BII contains a peptidase domain including a typical double β-barrel fold and previously unreported α-helical domain. The structures of peptide complexes revealed that the α-helical domain covers the active-site cleft and the side chain of Asn330 in the domain forms hydrogen bonds with the N-terminus of the bound peptide. These observations indicate that the α-helical domain regulates the exopeptidase activity of DAP BII. Because S46 peptidases are not found in mammals, we expect that our study will be useful for the design of specific inhibitors of S46 peptidases from pathogens.
- 1] School of Pharmacy, Iwate Medical University, 2-1-1 Nishitokuta, Yahaba, Iwate 028-3694, JAPAN [2].
Organizational Affiliation: