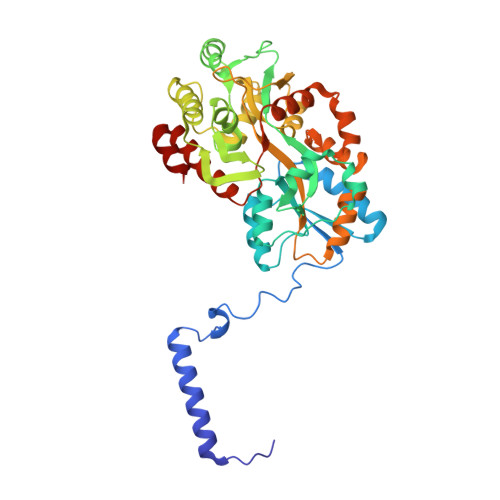
Co-translational folding of alpha-helical proteins: structural studies of intermediate-length variants of the lambda repressor.
Hanazono, Y., Takeda, K., Miki, K.(2018) FEBS Open Bio 8: 1312-1321
- PubMed: 30087834
- DOI: https://doi.org/10.1002/2211-5463.12480
- Primary Citation of Related Structures:
3WOA - PubMed Abstract:
Nascent polypeptide chains fold cotranslationally, but the atomic-level details of this process remain unknown. Here, we report crystallographic, de novo modeling, and spectroscopic studies of intermediate-length variants of the λ repressor N-terminal domain. Although the ranges of helical regions of the half-length variant were almost identical to those of the full-length protein, the relative orientations of these helices in the intermediate-length variants differed. Our results suggest that cotranslational folding of the λ repressor initially forms a helical structure with a transient conformation, as in the case of a molten globule state. This conformation subsequently matures during the course of protein synthesis. Structural data are available in the PDB under the accession numbers http://www.rcsb.org/pdb/search/structidSearch.do?structureId=5ZCA and http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3WOA.
- Department of Chemistry Graduate School of Science Kyoto University Japan.
Organizational Affiliation: