A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
Fukumoto, K., Onoda, A., Mizohata, E., Bocola, M., Inoue, T., Schwaneberg, U., Hayashi, T.(2014) ChemCatChem
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2014) ChemCatChem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UPF0678 fatty acid-binding protein-like protein At1g79260 | 174 | Arabidopsis thaliana | Mutation(s): 5 Gene Names: At1g79260, At1g79260.1, YUP8H12R.14 EC: 5.99 | 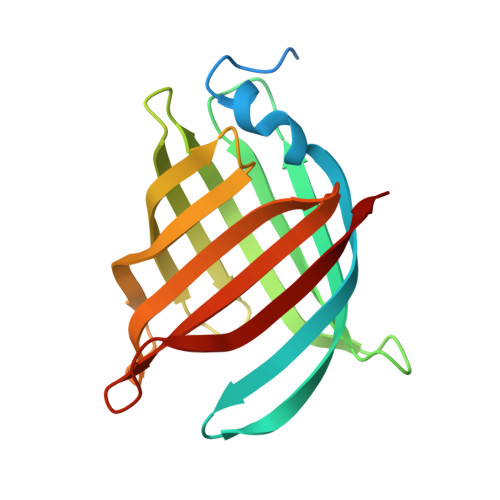 | |
UniProt | |||||
Find proteins for O64527 (Arabidopsis thaliana) Explore O64527 Go to UniProtKB: O64527 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O64527 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | B [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.94 | α = 90 |
| b = 79.41 | β = 90 |
| c = 36.477 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BSS | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |