Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Kanai, R., Ogawa, H., Vilsen, B., Cornelius, F., Toyoshima, C.(2013) Nature 502: 201-206
Experimental Data Snapshot
(2013) Nature 502: 201-206
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium/potassium-transporting ATPase subunit alpha-1 | A, D [auth C] | 1,016 | Sus scrofa | Mutation(s): 0 EC: 3.6.3.9 (PDB Primary Data), 7.2.2.13 (UniProt) Membrane Entity: Yes | 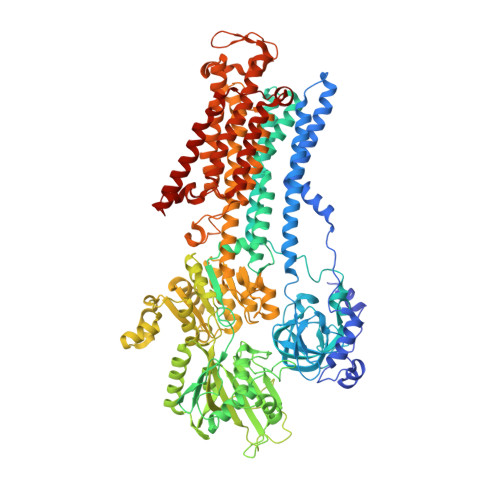 |
UniProt | |||||
Find proteins for P05024 (Sus scrofa) Explore P05024 Go to UniProtKB: P05024 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05024 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium/potassium-transporting ATPase subunit beta-1 | B, E [auth D] | 303 | Sus scrofa | Mutation(s): 0 Membrane Entity: Yes | 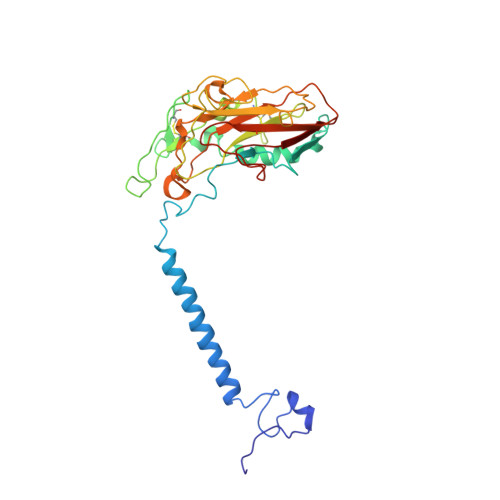 |
UniProt | |||||
Find proteins for P05027 (Sus scrofa) Explore P05027 Go to UniProtKB: P05027 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05027 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na+/K+ ATPase gamma subunit transcript variant a | C [auth G], F [auth E] | 65 | Sus scrofa | Mutation(s): 0 Membrane Entity: Yes | 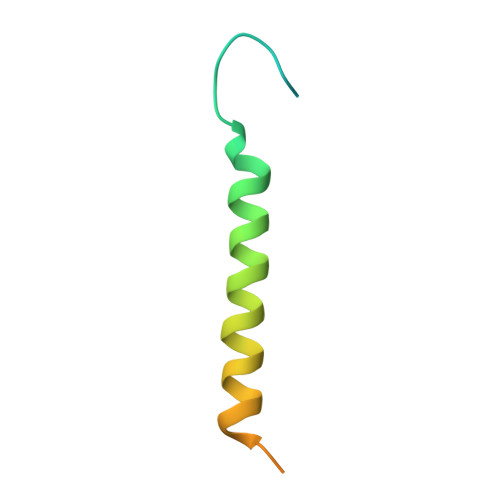 |
UniProt | |||||
Find proteins for Q58K79 (Sus scrofa) Explore Q58K79 Go to UniProtKB: Q58K79 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q58K79 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EFO Query on EFO | KA [auth C], U [auth A] | Oligomycin A C45 H74 O11 MNULEGDCPYONBU-AWJDAWNUSA-N |  | ||
| PC1 Query on PC1 | GA [auth C] HA [auth C] IA [auth C] JA [auth C] NA [auth D] | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H88 N O8 P NRJAVPSFFCBXDT-HUESYALOSA-N |  | ||
| ADP Query on ADP | BA [auth C], J [auth A] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| CLR Query on CLR | LA [auth D] MA [auth D] O [auth A] P [auth A] PA [auth E] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| NAG Query on NAG | OA [auth D], W [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| ALF Query on ALF | H [auth A], Z [auth C] | TETRAFLUOROALUMINATE ION Al F4 UYOMQIYKOOHAMK-UHFFFAOYSA-J |  | ||
| MG Query on MG | AA [auth C], G [auth A], I [auth A], Y [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | CA [auth C] DA [auth C] EA [auth C] FA [auth C] K [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 106.285 | α = 90 |
| b = 210.183 | β = 90 |
| c = 256.086 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BSS | data collection |
| PHASER | phasing |
| PHENIX | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |